Can You Sequence an Antibody?
What does it mean to sequence an antibody?
Antibodies are tetrameric proteins comprised of two identical heavy chains and two identical light chains. One naturally occurring exception to this is the heavy chain-only antibodies produced by camelids and cartilaginous fish. For a crash course in antibodies, check out our Antibody Reference Guide and our Nanobody Cheat Sheet.
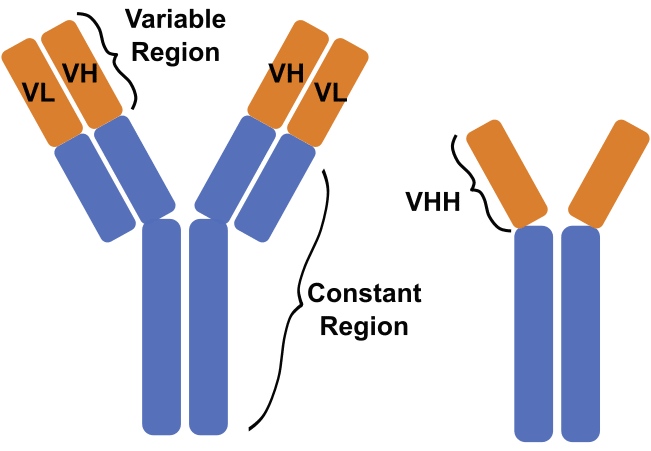
Antibody structure for the tetrameric conventional antibody (left), and the dimeric heavy chain-only antibody (right).
To sequence an antibody means to recover the primary amino acid sequence for the heavy chain and the light chain. In some cases, sequencing the entire antibody from N-term to C-term of both chains is needed. In other cases, antibody sequencing may target just the variable regions, or even just the complementarity-determining regions (CDRs).
In conventional IgGs, the heavy chain (including the variable region and constant region) is approximately 440 amino acids long, while the light chain is about half that length at 220 amino acids long. There can be considerable variability across isotypes and across species.
Why would you sequence an antibody?
Determining the sequence of an antibody is a requirement for many downstream activities.
-
Intellectual Property: In order to patent an antibody, the primary sequence is needed.
-
Antibody Optimization: In order to optimize the antibody, the primary sequence acts as a starting point for affinity maturation, humanization, or isotype switching.
-
Recombinant Production: Antibody genes can be synthesized and expressed heterologously in cells. Producing antibodies recombinantly ensures batch to batch consistency and an endless supply.
How do you sequence an antibody?
The method to sequence an antibody depends on what the status/form of the antibody. They key questions to ask about the sample are:
- Does the sample contain antibody protein or does it contain antibody-producing cells?
- Does it contain one clonal populational of antibodies (monoclonal) or is expected to be a mix of different antibodies (polyclonal)?
Antibody protein sequencing
Protein sequencing via mass spectrometry has become the most common method for sequencing antibody proteins.
Monoclonal antibody sequencing
A monoclonal antibody consists of many protein molecules encoding the same sequence. Monoclonal antibodies are the product of populations of antibody secreting cells such as hybridomas or transient production of recombinant antibodies.
De novo monoclonal antibody sequencing methods, like our own Valens, enzymatically digest the antibody into peptides (typically 15-30 amino acids long) which are easier to sequence using mass spectrometry than larger molecules. Valens uses multiple enzymes that cleave the antibody at different locations to ensure many overlapping peptides are generated. Once the peptides are sequenced, they are assembled back into a full-length heavy chain and/or light chain sequence.
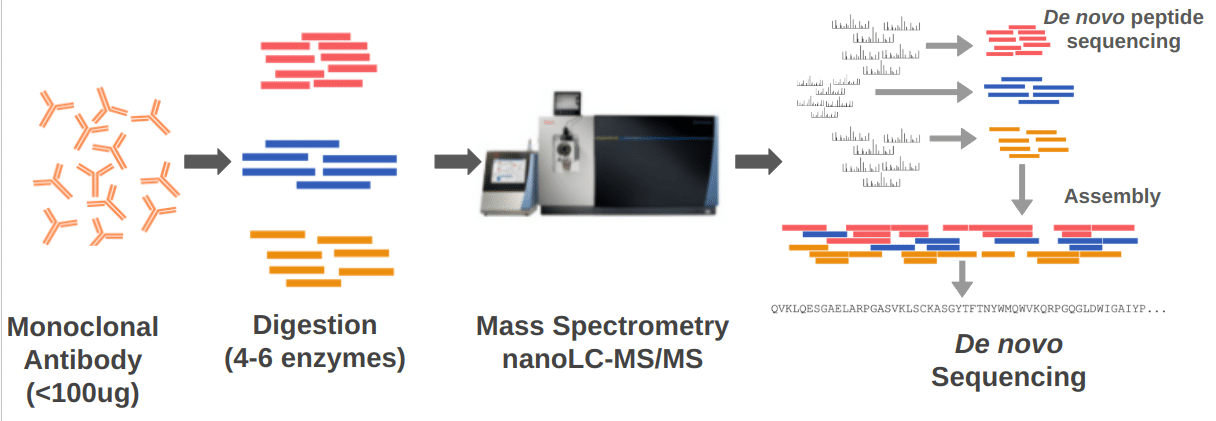
The mass spectrometry process for monoclonal antibody sequence methods, like Valens, is shown above. The antibody is digested with multiple enzymes into short peptides. The peptides are analyzed by LC-MS/MS. The Valens software performs de novo peptide sequencing and then assembles the peptides into a full-length antibody.
Polyclonal antibody sequencing
When an antibody is the product of a complex mixture of antibody-secreting cells and contains multiple antibody clones it is called a polyclonal antibody. Most polyclonal antibodies are derived from animals including human patients. They can also be generated by pools of recombinantly generated antibody secreting cells or hybridoma libraries, though this is less common.
Polyclonal antibodies can vary widely in their complexity, from mixtures of monoclonal antibodies to natural immune responses to vaccination or infection. Mass spectrometry is the preferred tool for polyclonal antibody sequencing, including our tool Griffin. However, given the high level of sequence homology between individual antibodies in a polyclonal mixture, the sample preparation, mass spectrometry analysis, and data analysis are all more complicated than for monoclonal antibodies. You can read more about the differences between monoclonal and polyclonal antibody sequencing in a recent blog post.

Clonality assessment is the first step in polyclonal antibody sequencing with Griffin. A network is created from the CDR3 sequences identified in each sample. A continuum of clonalities is shown above from a monoclonal antibody (with a modification variant) shown on the left to a natural human response to infection on the right.
Find out more about clonality assessment with Griffin in a recent blog post.
DNA or RNA-based antibody sequencing
An alternative to antibody protein sequencing is DNA or RNA-based antibody sequencing. For this process, antibody-encoding cells are needed in order to capture antibody RNA transcripts or genes. There are many cell types used for this process, as well as many technologies for sequencing. Our Reptor platform can be used on many input cell types including single B cells, hybridomas, phage libraries, splenocytes, peripheral blood mononuclear cells, lymph nodes, and more! Reptor targets the variable region of the antibody heavy chains and light chains to enrich for antibody sequences, and utilizes next-generation sequencing’s economies of scale.
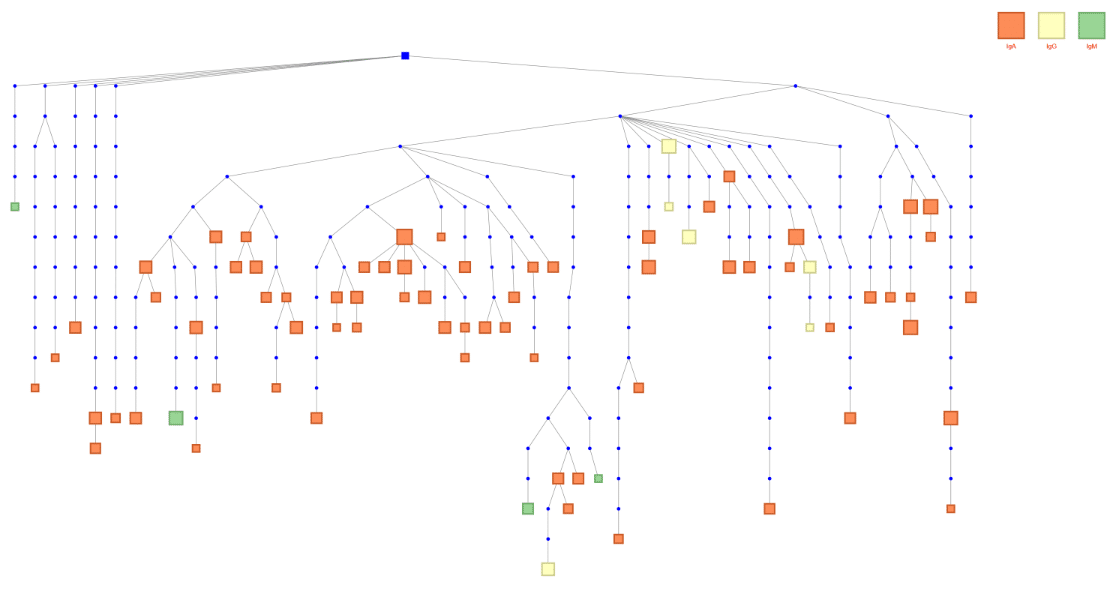
Visualization of an antibody clonal lineage that spans several antibody isotypes using Reptor.
What to do after sequencing
Once you have an antibody sequence there are many options for what comes next.
-
Recombinant expression: The antibody genes can be synthesized, cloned into an expression vector, and transiently produced in a variety of cell types.
-
Antibody engineering: The sequence of the antibody can be altered in silico prior to recombinant expression to change the isotype, or reformat as a single chain fragment. Mutant variants can be produced to find molecules with better characteristics than the original antibody.
-
Sequence search: The antibody sequence or sequence fragment can be used as a query to find similar sequences in public repositories, patent databases, or in antibody repertoires generated from the same campaign (a process called hit expansion).
-
Clone tracking: When sequencing multiple related samples, such as longitudinal studies in patients or across phage library panning rounds, an antibody sequence and its relatives can be identified and measured across samples.
