Case Study: Reptor Phage Enrichment
Project Summary
Phage display is a powerful toolkit for antibody discovery and engineering. In this study, we used phage display to identify Cas9-binding single domain (VHH) antibodies. In addition to traditional colony picking, we used our Reptor immune repertoire sequencing platform to monitor the phage panning process and identify additional hits that were not found in the traditional workflow.
Reptor is a proprietary antibody repertoire analysis workflow to enable deep interrogation of the immune response and antibody discovery processes. In this project, we used Reptor to analyze B cells from peripheral blood of an immunized llama as well as a phage library generated from the same bleeds.
Methods
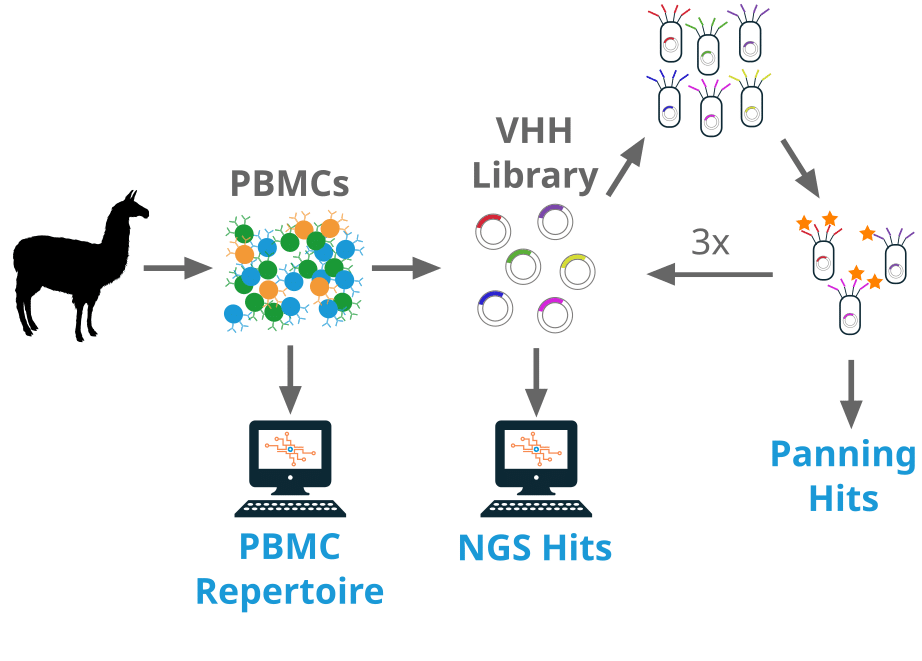
A llama was immunized with Cas9 and peripheral blood mononuclear cells (PBMCs) were collected. A VHH library was constructed from the PBMCs. The library went through 3 rounds of selection against the immunogen, Cas9. DNA was extracted from an aliquot of the pre-panned library (R0), and after each round of selection (R1, R2, and R3).
From the final R3 library, 88 transformants were found to be binders to the target, which after sequencing revealed 26 unique clones as defined by CDR3 sequence. These clones make up the ‘panning hits’.
In addition, Reptor was used to sequence the pre-panned library as well as the R1, R2, and R3 libraries. Enrichment analysis was performed to identify library sequences that increased in abundance between R0 and R3. These hits make up the ‘NGS hits’.
Finally, from the same PBMC samples used to create the phage library, Reptor constructed an antibody repertoire for the heavy chain only isotypes (IgG2B and IgG2C). This repertoire was used for hit expansion (‘PBMC repertoire’).
Results
PBMC versus Phage Repertoire Analysis: We used Reptor to compare the PBMC repertoire derived from PBMCs to the phage library repertoire. The PBMC repertoire contained 478,828 CDR3s, while the phage library (combining R0, R1, R2, R3) contained 169,447 clones. The majority of the clones found across all phage libraries (66%) are present in the R0 library, which is more diverse than any of the post-panning libraries. There is little overlap between the CDR3s found in the two repertoires with 44,066 of 604,209 (~7%) CDR3s shared between the phage libraries and the PBMC repertoire.
Phage Enrichment Analysis: In total 1,512 antibodies were identified in all panning rounds. From these, the enrichment score after each panning round versus the pre-panned library were calculated and plotted.
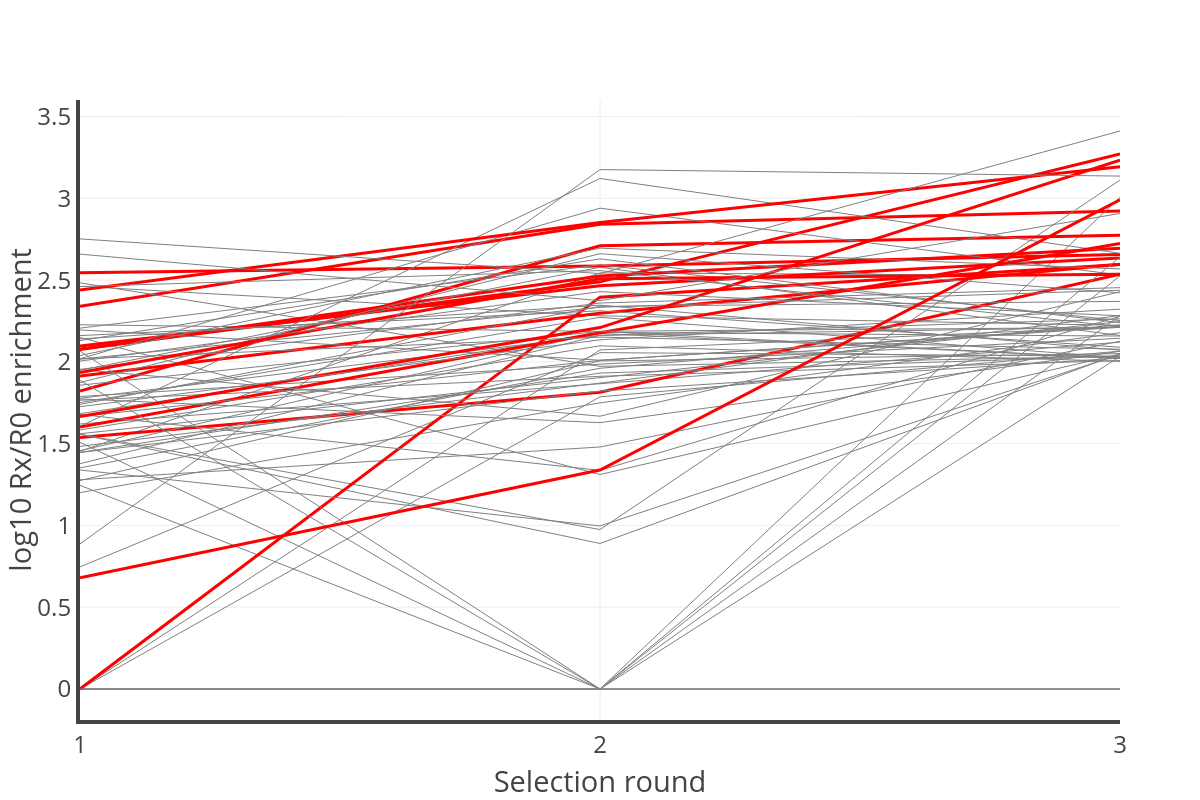
A plot of enrichment of clones after each round of panning (1, 2, or 3) versus the pre-panned library. The red highlighted lines are the 12 VHHs selected for testing.
Binding analysis: From the 25 sequences with final enrichment score of at least 2.5 between R3 and R0, 12 with the highest enrichment slopes were selected for testing. Of the 12 NGS hits tested, all 12 showed binding to the target in ELISA. The 12 NGS hits had distinct CDR3s from the panning hits. Among the 26 panning hits tested, 24 expressed but only 6 showed binding to the target.
Hit expansion: All 12 of the NGS hits have relatives in the PBMC-derived repertoires. Reptor revealed between 2 and 78 sequences that share the same CDR3 as the NGS hits.
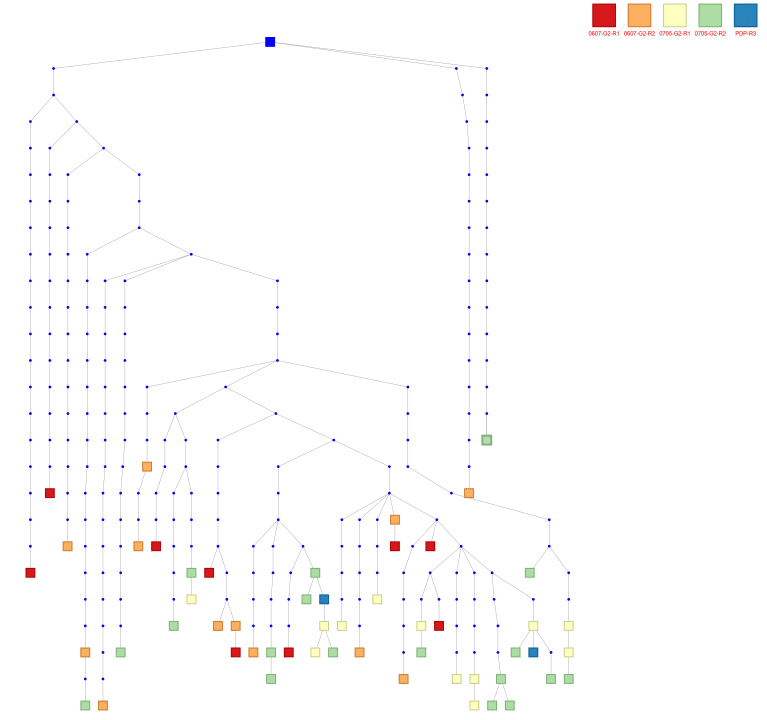
A lineage tree of sequences from the PBMC-derived repertoire that share CDR3 and germline V gene with one of the NGS hits. Red and orange nodes are sequences derived from early bleeds while yellow and green nodes are sequences derived from late bleeds. The two blue nodes are from the NGS hits.
Reptor uses hit expansion to find related hits with different characteristics from the primary hit. For example, in the lineage above, 3 of the sequences contain N-glycan motifs in the CDR2. Hit expansion identifies alternate sequences with identical CDR3 but without the N-glycan motif.
Conclusion
- Reptor analysis of phage libraries reveals novel clones that are enriched across panning, but not identified due to undersampling at end.
- NGS enables hit expansion, with all enrichment hits having relatives the remnant PBMCs.
Relevant links:
Related Posts
VHH Antibody Repertoire to Phage Library Comparison
Project Summary Camelids such as llamas and alpacas produce conventional antibodies as well as heavy-chain only antibodies (HCAbs) from which the small, stable antigen-binding domain (VHH) can be extracted. Abterra Biosciences developed Reptor, a proprietary antibody...
Case Study: Reptor Biological vs Technical Replicates
Project Summary Next-generation sequencing of B cell populations (Rep-Seq) has become a powerful tool in immunology and antibody discovery. At Abterra Bio we developed Reptor, an integrated Rep-Seq sequencing and analysis service. A key goal of Reptor is to provide...
New Service: Alicanto for Antibody Discovery from Human Serum
We recently had the pleasure of attending the PEGS Boston Conference. We had the privilege of showcasing our work through a poster presentation, a talk, and an exhibit booth. Among the highlights were engaging with experts, exchanging ideas, and strengthening our...