VHH Antibody Repertoire to Phage Library Comparison
Project Summary
Camelids such as llamas and alpacas produce conventional antibodies as well as heavy-chain only antibodies (HCAbs) from which the small, stable antigen-binding domain (VHH) can be extracted. Abterra Biosciences developed Reptor, a proprietary antibody repertoire analysis workflow that can be used to analyze both natural repertoires from peripheral blood mononuclear cells (PBMCs) and phage libraries. In this project we used Reptor to compare the VHH antibody repertoire characteristics across these two sources from an immunized llama. Reptor’s interactive output report automatically generates presentation-ready (or blog-ready) images.
The camelid IGH locus rearranges into both conventional antibodies with a heavy chain and a light chain (IgG1), as well as the heavy-chain only antibodies. The V genes of the locus are separated into two sets: those typically used by conventional antibodies (generally having a hydrophobic patch in framework 2 where the light chain associates), and those typically used by HCAbs (generally lacking the hydrophobic patch). The incorporation of the conventional antibody V genes has been previously reported in HCAbs.
Key questions we answer in this blog post:
- Do the general characteristics of the phage library match the PBMC samples taken from the same time point?
-
What is the clone level overlap between the phage library and PBMCs?
Data description
A single llama was immunized with a protein immunogen over the course of 14 weeks. PBMCs were collected at week 9 and week 13. The PBMCs were lysed and RNA was extracted. The RNA was aliquoted for use either in phage library construction or repertoire sequencing as shown in the overview figure below.
Reptor was used to analyze three repertoires; the phage library (pre-panning, the week 9 VHH antibody repertoire, and the week 13 VHH antibody repertoire.
There are key differences between the sequencing library generated from the phage library and those generated from the PBMCs directly.
- Primers for amplification: When cloning the VHH sequence into a phage vector, only the variable region is incorporated. Primers must anneal to regions inside the variable region, typically frameworks 1 and frameworks 4. In contrast, amplification for sequencing directly from the PBMCs can use primers outside of the variable region, typically in the 5′ UTR and hinge of the constant region. Using primers outside of the variable region avoids biasing the sequence composition and can provide isotype information since the hinge distinguishes the two HCAb isotypes (see more below).
- Targeted enrichment of HCAbs: Amplification of both conventional antibody heavy chains and HCAbs is a common first step of the VHH phage library construction process (Pardon et al 2014). The HCAb amplicons are identified based on size in a gel and then undergo a second round of amplification using nested primers that anneal to the variable region. Size-based separation may result in contamination of HCAbs with conventional antibodies, which would be indiscernible from true HCAbs after the second PCR.
Heavy-chain only antibodies in llamas are expressed as two isotypes with different hinge lengths: IgG2B (long hinge) and IgG2C (short hinge); also referred to as IgG2 and IgG3 respectively. From the phage library, it’s impossible to determine which isotype the VHH expressed due to the nested PCR described above. Therefore, for the phage library, the data is analyzed without this information. For the PBMC-derived repertoires we amplified and analyzed these isotypes separately. In this llama we found that ~10% of clones in the PBMC repertoire were expressed as IgG2C, suggesting that the IgG2B PBMC repertoire may be a closer match to the phage library.
During the construction of the phage library, primers that anneal within the variable region must be used. In contrast, the primers used for amplifying the VHHs from the PBMCs anneal outside of the variable region. This difference in primer sets can result in differences in VHHs based on germline gene usage. It may also cause overwriting of any framework 1 or framework 4 mutations in the phage library, as these will match the primer sequences.
Results
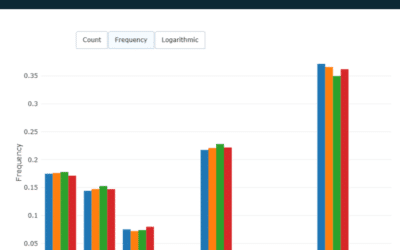
The phage library contained a range of CDR3 length that skewed slightly shorter than PBMC-derived repertoire.
The CDR3 length distribution for the week 9 IgG2B, week 13 IgG2B, and phage library repertoires.
The CDR3 length distribution is consistent across the two PBMC libraries, and closely matches the phage library distribution, however, the phage library is enriched for shorter CDR3s. The week 13 repertoire appears to be skewed slightly towards longer CDR3s.
The CDR3 length distribution for the week 9 IgG2C, week 13 IgG2C, and phage library repertoires.
The CDR3 length distributions between the phage library and IgG2C repertoires is quite distinct. The IgG2C repertoires overall appear to favor longer CDR3s than either the IgG2C repertoires or the phage library.
The phage library contains VHHs that are slightly less mutated from germline than the PBMC-derived repertoire.
The phage library and PBMC repertoires have similar mutation distributions, however, there is a slight preference towards less mutated sequences in the phage library.
The histogram of V gene amino acid mutations for the phage library and the PBMC repertoires for IgG2C.
Similar to what we’ve observed previously, IgG2C repertoires tend to be more mutated than IgG2B, and consequently the phage library is less mutated than the IgG2C repertoire.
One explanation for the lower mutation rate may be that the primers used to construct the phage library suppress mutations in the framework 1 and framework 4. The plots on the right show the amino acid mutations of the PBMC repertoire compared to the phage repertoire at the first 27 amino acids. Indeed, diversity of the phage library is lower for the first 9 amino acids, except for positions 1 and 2 which show a common mutation from QVQ to ELQ or EVQ.
Amino acid mutations by position for the first 27 amino acids of the VHHs from the week 13 IgG2B PBMC repertoire.
Amino acid mutations by position for the first 27 amino acids of the VHH from the phage library.
The phage library and IgG2B PBMC repertoires had similar germline V gene usage, however the phage library incorporated more conventional V genes.
The V gene usage among the HCAb germlines is comparable across the libraries, and exhibits focused usage of a small set of germlines. As described previously, this is typical of camelid repertoires.
Though accounting for a minority of the repertoire (<20% of VHHs), the phage library does incorporate more VHHs that use a conventional germline V gene than the background repertoires.
The phage library behaves like a biological replicate of the PBMC repertoires
CDR3 overlap across the three repertoires (IgG2B only for the PBMC repertoires) was consistent with overlap across biological replicates. The Jaccard similarity (CDR3s in the intersection over CDR3sin the union of the sets) ranged from 0.07 to 0.09). CDR3 overlap between the IgG2C repertoires and the phage library (not shown), was much lower due to the low total CDR3 count (Jaccard similarity of 0.01-0.02).
Take home message:
- Phage library construction can influence the diversity of the final library. Primers can overwrite framework mutations, and size-separation based methods may lead to inadvertently incorporating conventional heavy chain sequences.
- Despite diversity-limiting challenges, the phage library captures good diversity of the background PBMCs, acting like a biological replicate. A diversity-expanding strategy would be to generate multiple libraries from the different PBMC timepoints and then pool to create a larger more diverse library.
-
Related Posts
Case Study: Reptor Biological vs Technical Replicates
Project Summary Next-generation sequencing of B cell populations (Rep-Seq) has become a powerful tool in immunology and antibody discovery. At Abterra Bio we developed Reptor, an integrated Rep-Seq sequencing and analysis service. A key goal of Reptor is to provide...
New Service: Alicanto for Antibody Discovery from Human Serum
We recently had the pleasure of attending the PEGS Boston Conference. We had the privilege of showcasing our work through a poster presentation, a talk, and an exhibit booth. Among the highlights were engaging with experts, exchanging ideas, and strengthening our...
Case Study: Reptor Phage Enrichment
Project Summary Phage display is a powerful toolkit for antibody discovery and engineering. In this study, we used phage display to identify Cas9-binding single domain (VHH) antibodies. In addition to traditional colony picking, we used our Reptor immune repertoire...
Get In Touch