Llama Antibody Discovery from Serum
Custom llama antibody discovery using Alicanto®
- An approach that doesn’t use phage or yeast display
- Directly capture the natural diversity of the immune response
- Sequences for all hits
- Quick go or no-go decision upfront
- No more panning!
Get started on your project.
Key Benefits
More Diverse Hits
- By avoiding cloning into phage and panning, Alicanto® directly samples the natural diversity of the llama immune response. This translates to more unique hits for your project.
Higher Hit Rate
- We enrich for the desired llama antibodies at the earliest stages of the process, which allows us to avoid wasting time on antibodies that don’t have the desired specificity.
In Vivo Engineering
- We deliver all related llama sequences to validated hits which allows you to discover new candidates with potentially different profiles of affinity, immunogenicity, and sequence liabilities.
**We recently analyzed a naive llama repertoire and cover a little more of the biology of llama antibodies in a blog post.
Llama Antibody Discovery Process
Deliverables
Sequences
At least 10 diverse candidate sequences selected by Alicanto®.
Interactive report
An interactive, web-based report of the entire dataset to investigate individual llama antibodies, clonal families, and repertoire-wide features.
Expressed llama antibodies
1mg of up to 10 antibodies validated by ELISA.
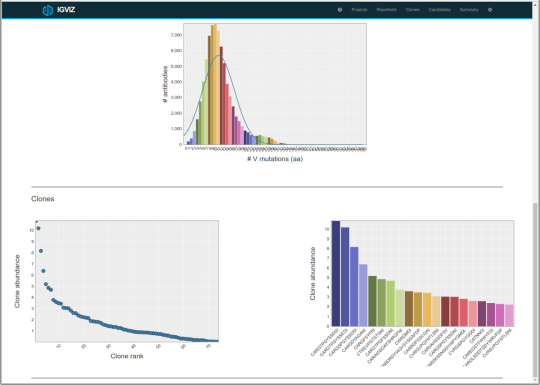
Other Alicanto® Services
Alicanto® uses mass spectrometry combined with next-generation sequencing to identify the highest affinity antibodies that have been selected by the native immune system. Our technology is flexible, and can be applied to a variety of host organisms, including rabbit, human, and llama.
Get In Touch
Tell Us About Your Project.
Need more information? We’re here to answer any questions you have.
Our Location
11588 Sorrento Valley Rd
Ste 16
San Diego, CA 92121
Publications by Abterra Bio founders and team members include:
- Safonova, et al. (2015). IgRepertoireConstructor: a novel algorithm for antibody repertoire construction and immunoproteogenomics analysis. Bioinformatics, 31, 12:i53-61.
- Cha, SW, Bonissone, S, Na, S, Pevzner, PA, Bafna, V (2017). The Antibody Repertoire of Colorectal Cancer. Mol Cell Proteomics, 16, 12:2111-2124.
- Bonissone, SR, Pevzner, PA (2016). Immunoglobulin Classification Using the Colored Antibody Graph. J Comput Biol, 23, 6:483-94.
