Single-molecule Ig Repertoire Sequencing from Lama glama

In recent years, camelids have become more than hearty travel companions due to their unconventional adaptive immune systems. Conventional IgG antibodies are tetrameric molecules comprised of two heavy chains and two light chains with molecular weights of roughly ~150 kDa. In addition to conventional antibodies, camelids also produce smaller homodimeric single chain antibodies, that have two heavy chains and no light chains. Since their discovery in the early 1990’s, scientists have found single variable domain antibodies to be better recognition tools than conventional antibodies for many detection, diagnostic, and therapeutic applications.
At Digital Proteomics, we’re profiling the llama and alpaca immune systems using our repertoire sequencing platform, Reptor™. We also perform custom single chain antibody discovery using our discovery technology, Alicanto®. To gain deeper insight into the camelid immune system, we employed Pacific Biosciences’ (PacBio) single molecule sequencing to generate a quantitatively accurate representation of IgG1 and IgG2 repertoires from a naive llama.
Tell Us About Your Project
Why single chain antibodies?
Easy to develop – The single variable domain (VHH) of single chain antibodies results in a much smaller 12-15 kDa recognition molecule than the comparable ~25kDa two variable domain single-chain variable fragments (scFv). Retaining similar affinity to whole antibodies, VHH have also been shown to be more stable and resistant to heat denaturation. Antibody production of VHH in yeast and bacterial cells is much simpler. scFv production requires linking of heavy and light chain variable domains for correct antibody conformation, whereas no linking is required for the single variable domain of VHH. Additionally, VHH’s small size allows for bacteria and yeast to produce the antibodies in higher yields.
Amenable to multi-specific formats – The next generation of cancer therapies favors bi-specific and multivalent antibodies, where a single molecule is capable of tethering, agonizing, or antagonizing multiple targets. For example, Blinatumomab redirects T-cells to tumor cells by targeting a tumor antigen (CD19) and a T cell marker (CD3). Multiple VHHs with different recognition targets can be engineered onto the same molecule, and correspondingly, has led to an increase in potential therapeutic molecules in clinical trials.
Faster tissue penetration – VHH enables much faster and higher contrast in-vivo tumor imaging. The small size allows for faster penetration and clearance of the recognition molecule. For example, SPECT imaging of small xenograft tumors with radionuclide labeled anti-EGFR VHH takes 45 min whereas imaging with radionuclide label cetuximab, a tetrameric antibody, takes at least 24 hours.
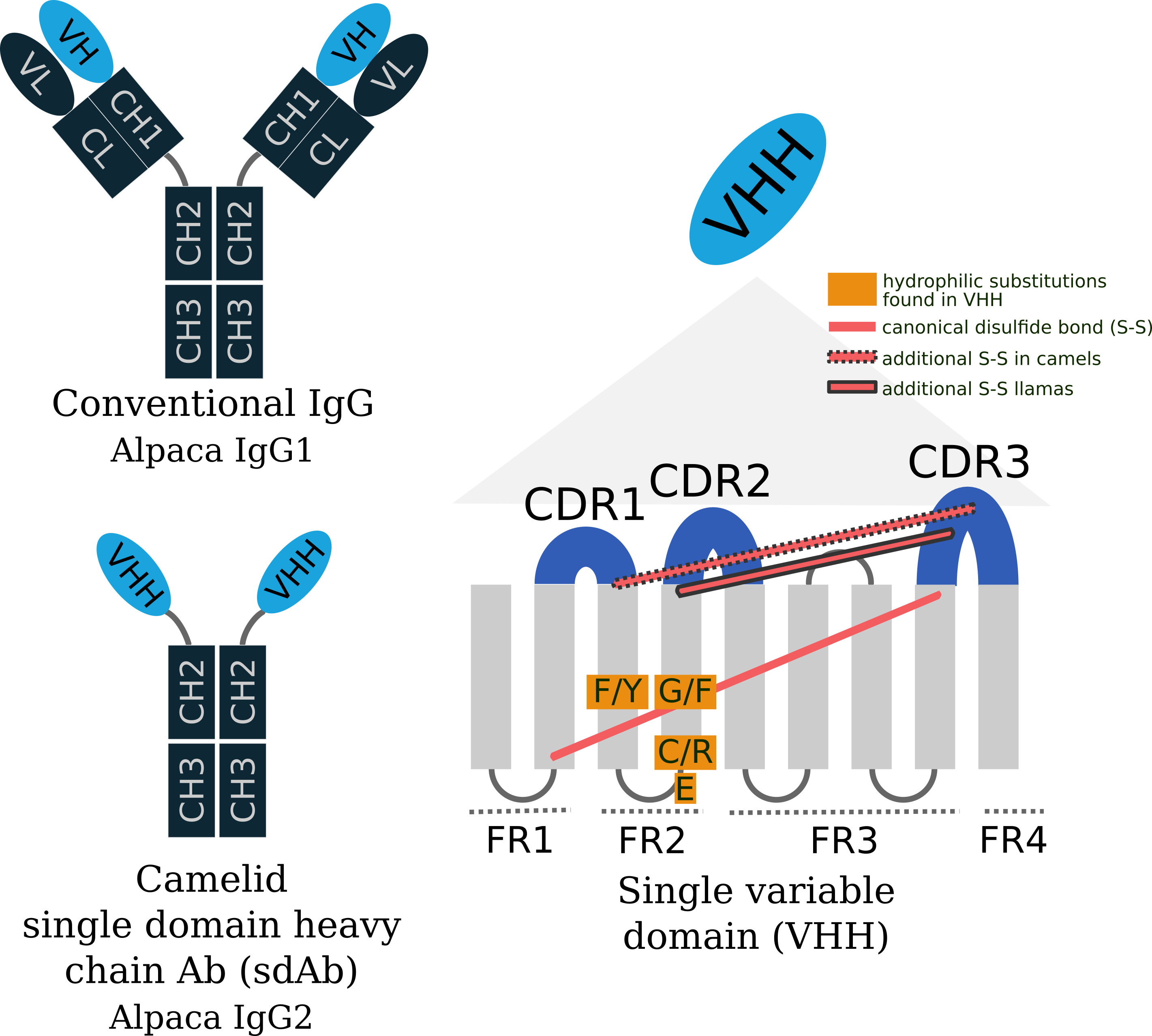
Single chain antibody structure
While the Lama glama IgH locus is yet to be fully characterized, the closely related Vicugna pacos (alpaca) locus has been, and is likely to be very similar to llamas. The Ig locus for alpaca is comprised of IgG1a, IgG1b, IgG2a, and IgG2b genes where VDJ recombination with IgG1 results in conventional antibodies and IgG2 result in single chain antibodies. Camelids reuse the same diversity and junction genes to produce both IgG1 and IgG2 antibodies, but have distinct VHH and VH variable genes. The locus contains 88 variable, 7 diversity, and 7 junction heavy chain genes. Of the 88 variable genes, there are 17 VHH genes all belonging to the variable domain heavy chain subgroup 3. A point mutation at the intron/exon splice junction of IgG2 prevents VHH from joining the CH1 domain. Without a CH1 domain, the heavy chain cannot form a disulfide bridge with CL domain of light chains, which occurs in conventional IgGs. Additionally, the framework 2 region of VHH genes contain several hydrophilic substitutions (Val → Phe or Tyr, Gly → Glu, Leu → Arg or Cys, Trp → Gly or Phe) as shown in the figure. Adding further structural stability, camels and llamas often have additional disulfide bonds between CDR3 and FR2 regions of VHHs.
Unbiased Ig repertoire sequencing from PBMC
We evaluated PacBio sequencing as the sample preparation introduces less amplification bias between IgGs and captures full-length antibody sequences. Most repertoire sequencing is performed on Illumina’s sequencing by synthesis instruments; however, the technology is limited to compare antibody sequences of less than 600bp. Single molecule sequencing is capable of producing longer sequences with the caveat of increased insertion and deletion errors. However, as antibody sequences for llama IgG1 and IgG2 are less than 2kbp, each antibody sequence is read multiple times and can be corrected to produce more accurate sequences. The corrected sequences are called circular consensus sequences (CCS) and constructed using a tool called Arrow.
Briefly, peripheral blood mononuclear cells (PBMC’s) from a naive llama were lysed, RNA was extracted, and enriched for IgG following a 5′ RACE protocol with a reverse primer located in CH2, which is shared by all IgG genes. The resulting DNA was prepared as a sequencing library and run on a PacBio Sequel System for 20 hrs. Our library generated 20.5 million subreads, which formed 440k CCS with every antibody sequence being read at least 3 times. These sequences were mapped to germlines IgG1a, IgG1b, IgG2b, and IgG2c. Antibody sequences were then VDJ labeled by aligning to V, D, and J gene references derived from other sequencing experiments. After labeling, CDR3’s were extracted from CCS and clustered into clones. Somatic hypermutations were called for each CCS with a V gene label.
Results
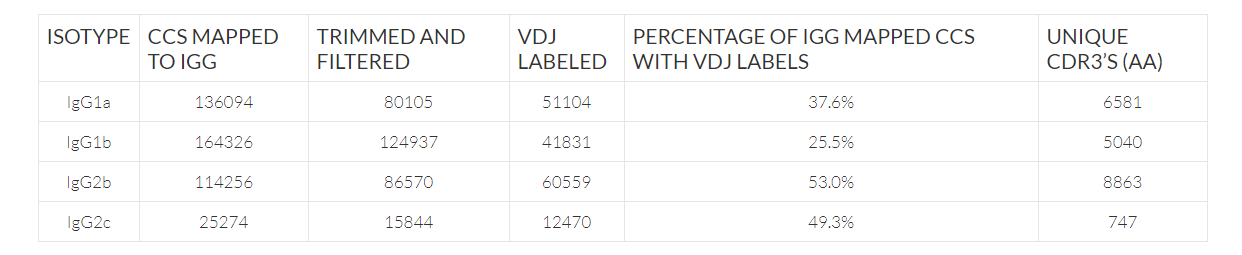
Like previous studies, VHH transcripts were less abundant compared to VH transcripts with 26.0% and 5.7% of CCS sequences of IgG2b and IgG2c types. However, after clustering antibody sequences into clones, IgG2b resulted in the most unique CDR3’s. This is likely due to an underestimation of unique CDR3’s for IgG1a and IgG1b, as opposed to a biologically significant increase in unique VHH CDR3’s.
Repertoire construction statistics by isotype
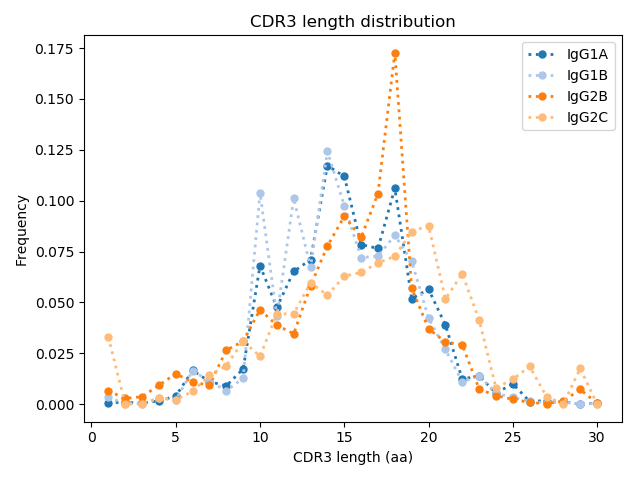
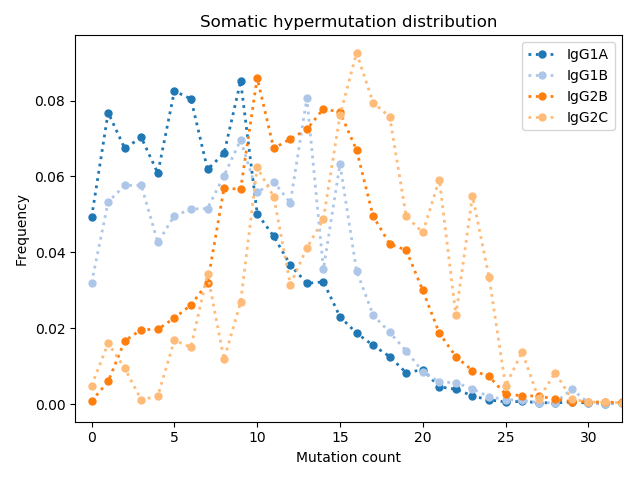
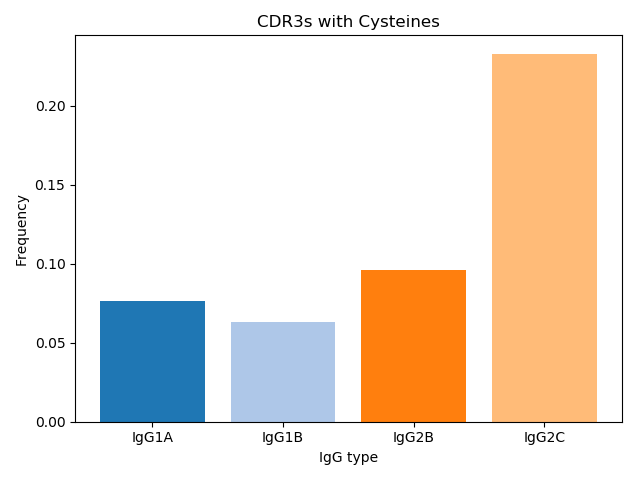
Unlike previous studies, an increase in VHH CDR3 length distribution was not observed. Li et al. 2016 have reported VHH has on average length of ~5 more amino acids than VH derived antibodies in camels. Similar to what was observed in camels, VHH antibodies in our naive llama had more somatic hypermutations than conventional antibodies. Others have reported VHH stability is ensured by an extra disulfide bond between framework 2 and CDR3. However, Li et al. reported that ~9% of both VH and VHH clones have cysteines in extracted CDR3’s. In our llama VHH repertoire, we observed a higher frequency of cysteines in CDR3’s. Surprisingly, 23.2% of IgG2c clones had CDR3’s with a cysteine.
Limitations of PacBio sequencing
Calling runs of the same nucleotide is challenging for PacBio sequencing. The IgG constant region genes are not expected to have mutations and each reference contained a cytosine homopolymer of length 5 or 6. We found ~20% of circular consensus sequences contained homopolymer errors at these cytosine sites.
Our repertoire analysis pipeline was developed for Illumina sequencing, which doesn’t explicitly model indel errors observed in single molecule sequencing. We saw a large attrition of antibody sequences at various quality filtering stages of our repertoire analysis pipeline, even when starting with minimum QV30 circular consensus sequences. Further specialization of repertoire construction and VDJ labeling may improve this attrition, however it is unlikely to significantly alter the somatic hypermutation profiles, and CDR3 properties shown above.
Tell Us About Your Project.
Need more information? We're here to answer any questions you have.
Related Links
Single-Domain Antibodies As Versatile Affinity Reagents for Analytical and Diagnostic Applications
Tetrameric and Homodimeric Camelid IgGs Originate from the Same IgH Locus
VHH antibodies: Emerging reagents for the analysis of environmental chemicals
Single-Domain Antibodies and the Promise of Modular Targeting in Cancer Imaging and Treatment