Polyclonal Antibody Sequencing in Camelids
Serum contains circulating antibodies with unique functional properties. This fundamental fact is why we choose to mine the serum over cells – despite being more challenging. Here we compare the two approaches for polyclonal antibody sequencing available at Abterra Bio by applying them to the problem of polyclonal antibody sequencing in camelids. Camelids, like llamas and alpacas, produce heavy chain-only antibodies devoid of light chains, and their variable domains can be produced independently as VHHs, or nanobodies.
Alicanto mines the serum with the aid of B cells, using a proteogenomic approach, while Griffin is informed exclusively by the serum proteomics. We recently compared these two polyclonal antibody sequencing methods, their benefits and limitations.
Alicanto searches a database of eligible antibody sequences, allowing for greater sensitivity due to the fact that peptide-spectrum matches (PSMs) can have less coverage and still be identified by relying on the prior information of the database sequences. While sensitive, only antibodies contained in the database can be identified with Alicanto. In immunoinformatics, however, databases are seldom comprehensive, and never complete leading to missed antibodies.
For Alicanto, the database is generated for each project using next-generation sequencing (NGS) of the B-cell repertoire. The database may be incomplete for a variety of different reasons:
- Biological: The B cell that created that particular serum antibody did not reside in the compartment where the B cell was captured, e.g., capturing PBMCs in blood when plasma cells in bone marrow produced the circulating antibodies, see Figure 1a for localization challenges for capturing cells that secrete antibodies (i.e., plasma cells), versus what is captured in the circulating PBMCs (i.e., memory B-cells).
- Statistical: The subsampling of the cellular repertoire did not capture the correct B cell with corresponding serum antibody. To illustrate this problem, Figure 1b shows the subsampling problem with B cells in humans – that you can only hope to capture ~10E-6 of the B cells (memory B cells).
- Technical: Other technical limitations to NGS that precluded identification and further reduce the total sampling, e.g., PCR errors, sequencing errors, transcript degradation, FDR sensitivity, etc.
This blog post will explore the differences in clones identified by proteogenomics (Alicanto) and fully de novo proteomics (Griffin) for polyclonal antibody sequencing in camelids to answer the question: how much of the serum antibody repertoire are we missing if we focus on B cells?
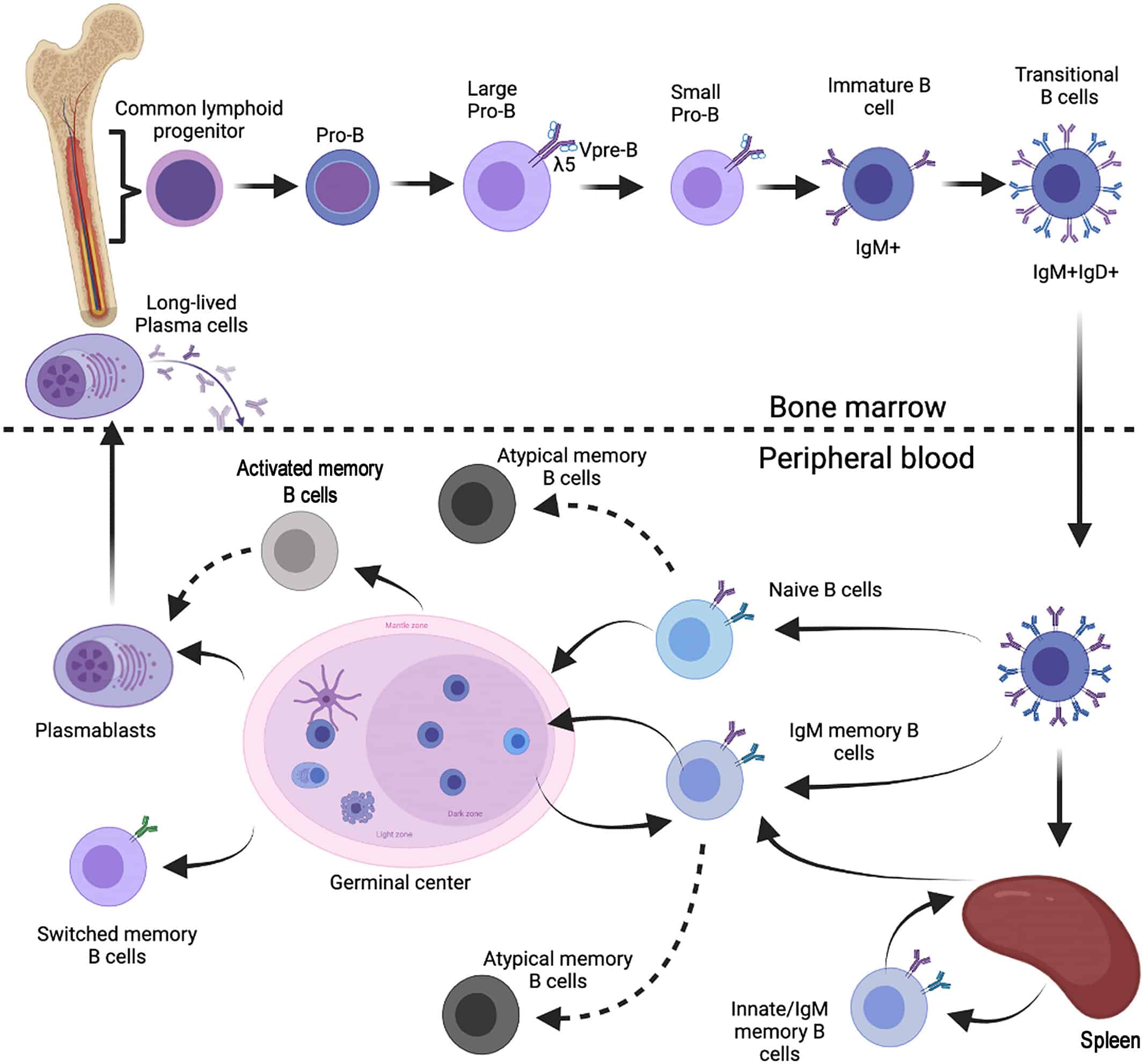
Figure 1a: Plasma cell and B-cell compartments compared to secreted antibodies. (Figure credit to Carsetti, Rita, et al. (2022)).
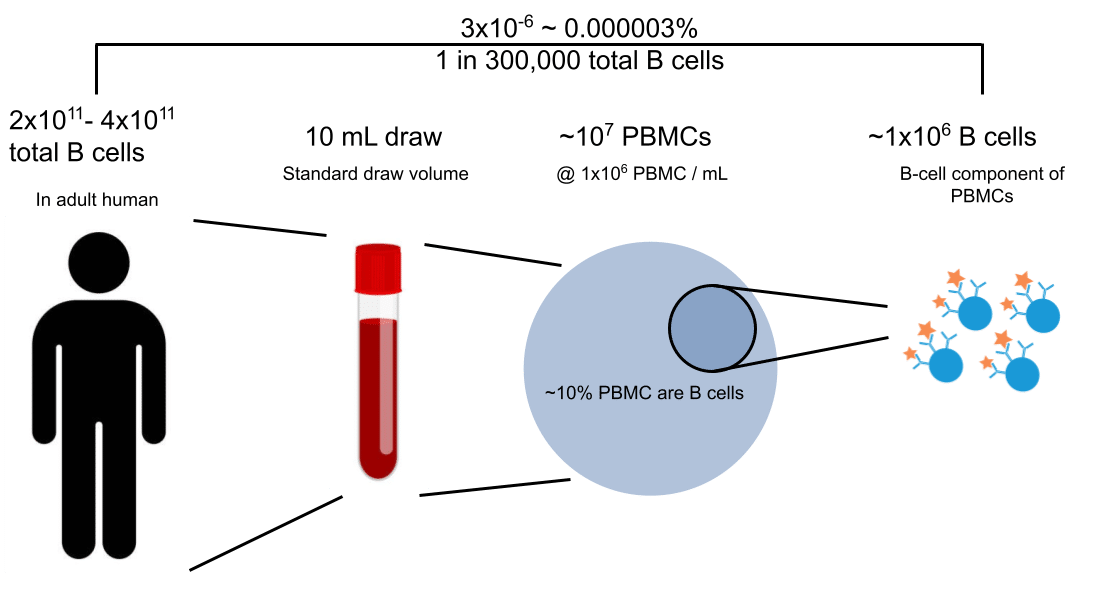
Figure 1b: Subsampling B cells from blood PBMCs scratches only the surface of the total ~3×10^11 B-cell population in humans (Sender et al., (2023)).
Background on polyclonal antibody sequencing in camelids
Given the two repertoires, serum and cellular, we have the following cartoon Venn diagram to illustrate the distinct categories of serum antibodies that can be detected with Alicanto versus Griffin.
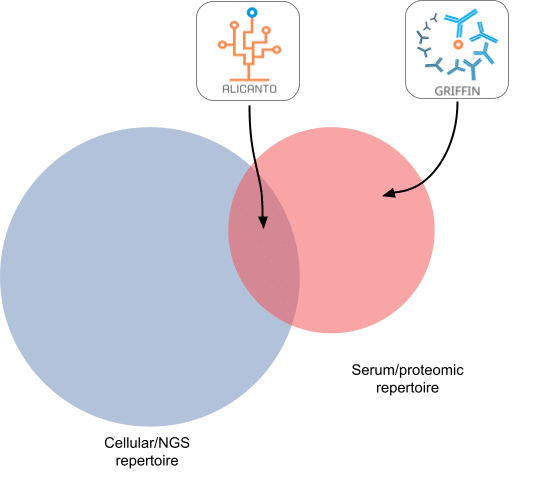
Figure 2: Alicanto vs Griffin as exemplified by cellular vs serum repertoire overlaps
The antibodies that overlap the serum and cellular repertoires are targeted by Alicanto (see Figure 2) – since it utilizes the cellular (NGS) repertoire to build a custom database for searching the serum-derived proteomics.
The entire serum compartment is mined by Griffin – relying on no cellular information for reconstruction. However, despite this, it is possible for Griffin to reconstruct clones/antibodies that are present in the cellular compartment, i.e., the intersection. The ratio of how many clones/antibodies that Griffin recovers that are in the intersection (and potentially accessible by Alicanto) versus only found in serum and exclusive to Griffin, is the question we seek to answer in this analysis.
Method
Utilizing an existing alpaca immunization, described in a previous blog post, we subjected the same polyclonal to mass-spectrometry for the purposes of sequencing with Griffin. Since this is a camelid project for VHH discovery, only CDR3 sequences from the heavy chain (CDRH3) will be considered. The similarity between Griffin and Alicanto clones is assessed at two different stages of granularity:
- Clonal assessment of CDRH3s
- Using fewer enzymatic digests, we capture full length CDRH3s and use this as Stage 1 clones. This represents a subset of the CDRH3s able to be fully captured by Griffin.
- As part of assembly we can recover additional CDRH3s that were not recovered in Stage 1. Some CDRH3s that were identified in Stage 1 will not have a corresponding full length contig.
- Assembly of full length contigs
- We can compare full length assemblies as candidates from Griffin with candidates from Alicanto.
- While unlikely to obtain exactly the same full-length contig, we can determine how similar each contig is within each similar/same clone.
Alicanto serum clones and candidates were derived from a prior run of the same polyclonal material, further discussion of that project is found on our previous post.
Results
Clonal comparison for polyclonal antibody sequencing in camelids
Griffin Stage 1 clones were clustered alongside the Alicanto-identified serum clones. CDRH3s are nodes, with edges connecting two nodes if they are within 80% sequence similarity. Nodes are colored as belonging to either Griffin (red), Alicanto (blue), or found in both (purple). For the purposes of this clustering, we treat Ile/Leu residues from Alicanto sequences as interchangeable and equivalent for computation of similarity. We find:
-
44 Griffin clones (Red)
-
146 Alicanto clones (Blue)
-
2 overlapping clones (Purple)
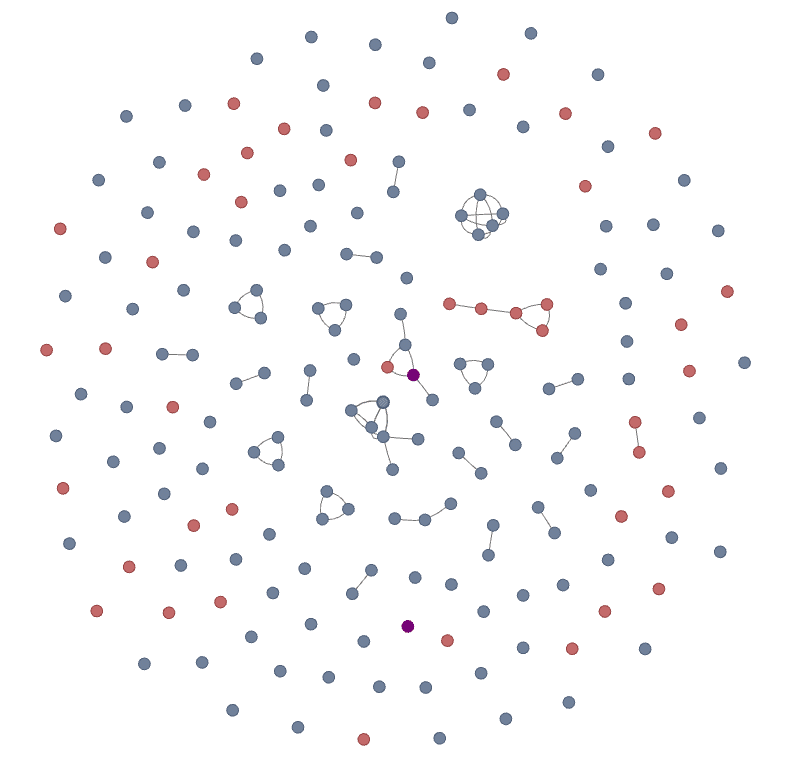
Figure 3a: CDR3 network of clones identified with Alicanto and Griffin. Each node is a distinct CDR3 amino acid sequence and edges are drawn between nodes that are within 80% sequence similarity.
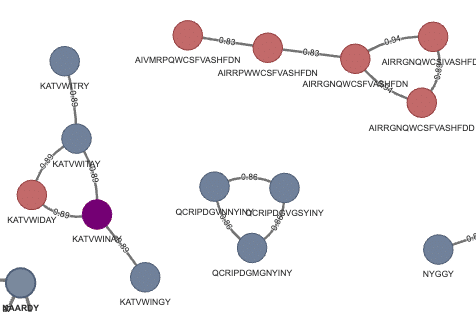
Figure 3b: Zoomed section with one overlapping clone (purple) between Griffin and Alicanto.
The two CDRH3s that were found in both Alicanto and Griffin were: KATVWINAY (seen in Figure 3a and 3b above), and ARGYISNYDG which is a singleton node in the network (seen in Figure 3a above).
The shared clone node, KATVWINAY, contained a validated binder from Alicanto derived CDRH3 KATVWLNAY (Ile → Leu), which was among the stronger binders of the validated candidates for this project.
Most of the CDRH3s found by Griffin are disjoint from the CDRH3s from Alicanto, with a few exceptions. One such exception is shown in Figure 3b, which highlights a clone cluster with one Griffin-specific, 3 Alicanto-specific, and one shared clone. All other clone clusters at this normalized 80% similarity threshold clustered within each method.
Since most clones in Griffin are not found exactly in Alicanto, we wanted to quantify what the most similar Alicanto clone would be for every Griffin clone. The histogram of maximum similarity for each Griffin clone is shown in the histogram in Figure 4:
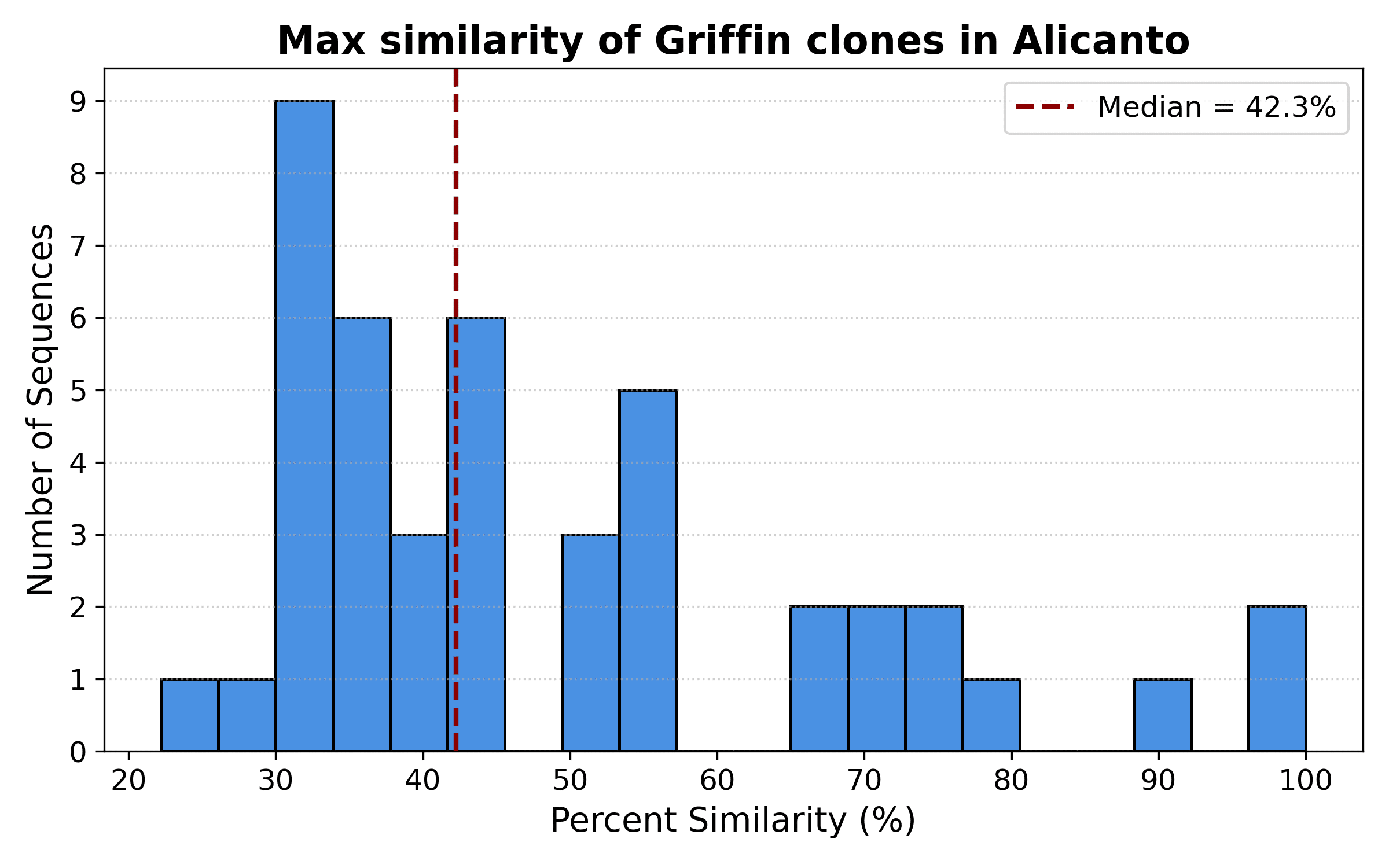
Figure 4: Similarity of closest Alicanto clone for each Griffin clone
There are a small number of clones above 80% similarity, as observed in the network in Figure 3a above, but most Griffin clones are no more than 43% similar to their closest Alicanto neighbor. This highlights how different clones found by Griffin in the serum.
Candidate comparison
For one of the two clones found identically in Alicanto and Griffin, KATVWLNAY, we wanted to see the differences in full length antibody sequences produced by transcript sequencing vs proteomic assembly. As part of assembly, the Ile from the CDR3 was converted to a Leu based on CLIP predictions. Figure 5 shows the multiple sequence alignment, highlighting shared and divergent mutations from germline V region IGHV3S53.
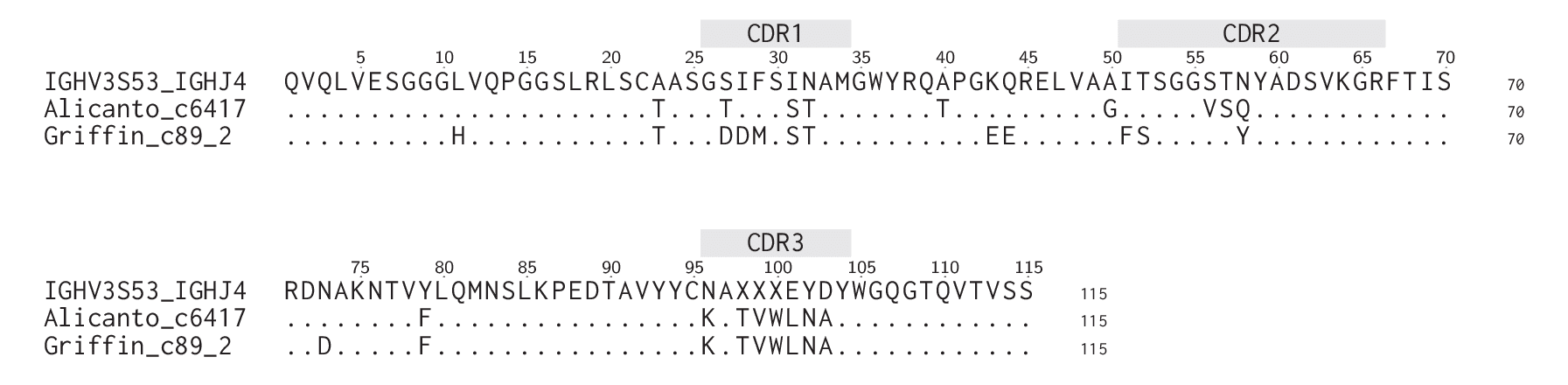
Figure 5: Alignments between Alicanto and Griffin candidates of clone KATVWLNAY
While both of these candidates are validated binders from the same proteomically identified clone, we see differences in upstream features of the antibody. It is notable that CDR1 has two mutations, I31S ad N32T, in common with Alicanto and Griffin candidates, while the Alicanto candidate has one additional mutation, and the Griffin candidate has three. CDR2s have three mutations each, with none overlapping between Alicanto and Griffin candidates.
Conclusion
Griffin and Alicanto are able to mine different parts of the serum polyclonal response; Alicanto sensitively captures the intersection of cellular and serum repertoire, while Griffin can capture the more abundant antibodies regardless of the cellular repertoire.
Interestingly, we found:
-
Most clones between Alicanto and Griffin are largely disjoint. There were 2/44 (4.5%) clones that overlapped
-
Alicanto identifies more total candidates due to it’s increased sensitivity
-
Griffin identifies candidates that are not found in the B cell repertoire.
-
-
Not only are Alicanto and Griffin clones mostly disjoint, they are very different – with most Griffin CDRH3s less than 43% similar to closest Alicanto CDRH3.
-
Full length sequences for one clone identify some of the same variants in CDR1 and FWR1/3
-
Different CDR2 mutations, and some FWR2 variants
-
Griffin can identify additional serum candidates not present in the B cell repertoire that is used by Alicanto and other discovery methods.
