VHH De Novo Antibody Sequencing
VHHs are 15kDa proteins that represent the antigen binding portion of heavy chain-only antibodies (HCAb) from llamas and alpacas. Their small size and stability make them ideal components of therapeutics such as bispecific antibodies or CAR-T therapies. In addition, VHHs can be expressed in non-mammalian expression systems such as E. coli, achieving excellent economies of scale during recombinant expression.
VHH de novo antibody sequencing is the process of identifying individual VHH sequences from a complex mixture, such as a polyclonal antibody (pAb). Our Griffin platform enables de novo antibody sequencing of pAbs from any species, including HCAbs from llamas and alpacas. There are many benefits to VHH de novo antibody sequencing. In this case study, we sequence a polyclonal antibody to convert it into a recombinant VHH that can be produced consistently, and indefinitely.
A llama was immunized against a protein antigen for the purpose of generating a polyclonal VHH antibody reagent. The animal died, preventing any further production of the pAb. The goal of the project was to identify monoclonal VHHs that recapitulate the pAb activity.
Mass Spectrometry Data Generation
The heavy chain-only antibodies were purified from total IgG from llama serum, and subsequently purified against the antigen using affinity chromatography. The pAb was digested with multiple enzymes to obtain peptide sequences that covered every region of the antibody. Using proprietary methods, long peptides were generated to enable assembly and phasing of CDRs.
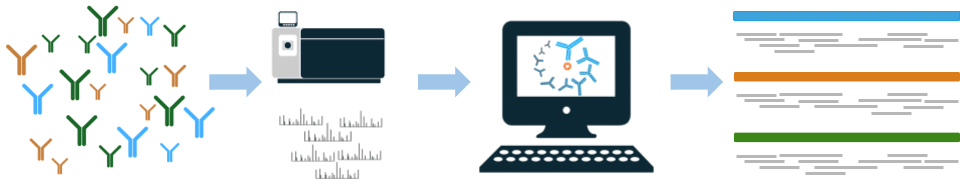
Figure 1: Overview for VHH de novo antibody sequencing with Griffin. The VHH polyclonal antibody is analyzed by LC-MS/MS. De novo peptide sequences are determined by Griffin, and assembled into full-length VHHs.
Griffin Stage 1: Clonality Assessment
A first-pass assessment of sequence diversity reveals the level of complexity of the sample. Peptide sequences are determined from the mass spectrometry data using in-house software. CDR3 sequences are extracted from the peptides to determine the complexity of the sample.
The number of CDR3 sequences and their similarity to one another are used to determine the clonal complexity.
A CDR3 network is constructed to evaluate similarity. Each CDR3 is represented by a node in the network, and an edge between nodes is added if the CDR3s are within an edit distance threshold (typically 80% similarity). The CDR3 network to the right shows two major families of CDR3s present.

Figure 2: An example CDR3 network for a polyclonal antibody shows two families of related clones.
Griffin Stage 2: Assembly and Sequencing
A deep second-pass analysis generates candidate assemblies. Short peptides provide information about each region of the antibody, while long peptides connect distal regions, such as CDRs. Each assembly is reviewed in silico to eliminate unnatural sequences and structures.
The figure shows the peptide support for one assembled VHH.

Figure 3: Peptides mapped to an assembled VHH sequence. The CDRs are noted as a track under the VHH sequence. Each colored bar below the sequence is a peptide identified from the mass spectrometry data. The colors represent different enzymatic digests and mass spectrometry runs. Peptides cover the entire VHH sequence, with many overlapping peptides providing support for the assembly.
Griffin Stage 3: Expression and Validation
CDR3 clustering at 80% similarity, single linkage, revealed 8 clone families. Representatives from each family were recombinantly expressed and tested. ELISA binding showed that 44% of all VHHs bound the target, and 5 of the 8 families showed activity. The 5 largest families all contained at least 1 binder, while the 3 singleton families all failed to show binding
A post-hoc analysis using molecular dynamics (MD) simulations of clones with similar sequences was predictive of failure to bind.

Figure 4: Molecular dynamics (MD) simulations identify unstable VHH sequences that are less likely to be functional.
Conclusion
VHH de novo antibody sequencing is possible with Griffin. In this case study we were able to identify individual VHHs that perform the same function as the starting pAb.
Rabbit pAb Sequencing
Griffin is species-agnostic, and can be used to sequencing polyclonal antibodies from other species – including rabbit like we did in a recent case study.
VHH Antibody Discovery
B cells and serum antibodies provide complementary information about the immune response. Our Alicanto platform combines B cell repertoire sequencing and serum antibody analysis to deliver diverse, functional VHHs.
Griffin vs Alicanto
What’s the different between serum-only antibody sequencing with Griffin and proteogenomic antibody sequencing with Alicanto? We investigate the advantages and trade offs of each approach in this blog post.



