Alicanto Antibody Discovery – Diversity
Alicanto® is Abterra Biosciences’ proprietary antibody discovery platform that mines the entire immune response to identify diverse, functional antibodies. For an overview of the proteogenomic process, read more about Alicanto®.
Alicanto benefits include:
- Full sequence information for candidate antibodies selected by Alicanto®
- Verifiable diversity in an interactive HTML report with data visualization.
- Transparency through a written summary of the Alicanto® process and findings
Alicanto antibody candidate sequences
The primary deliverable of Alicanto® is a set of candidate sequences that have the desired function. Candidates are selected from the repertoire using Alicanto’s proprietary machine learning model that incorporates information from both the genomics and proteomics data. The exact number of candidates is determined based on the stringency of the functional requirements.
The candidate sequences are provided in a tab-delimited format developed by the Adaptive Immune Receptor Repertoire (AIRR) community. Included in the candidate sequence file are:
-
-
- Nucleic acid sequence of variable region
- Amino acid sequence of variable region
- Complementarity-determining region (CDR) sequences
- Germline V, D, and J gene calls
- Isotype/locus
-
Tell Us About Your Project
Alicanto® interactive report
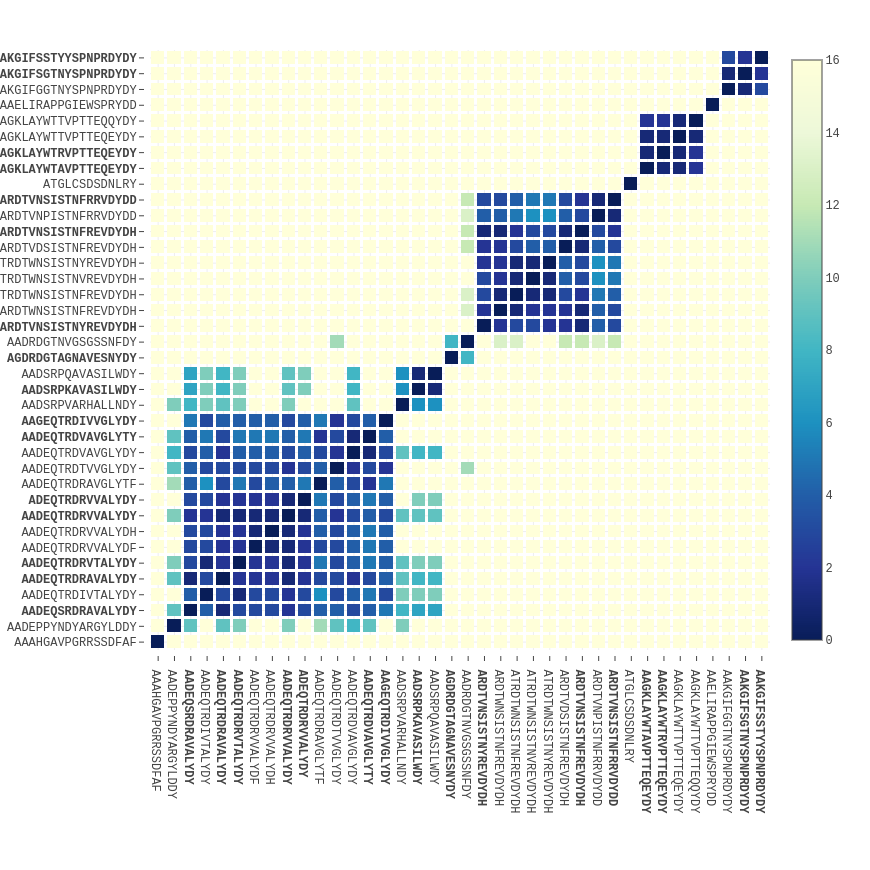
CDR3 families are detected by proteomics
Through the process of affinity maturation, the antibody response to immunization produces families of related antibodies. Alicanto® can detect the presence of multiple family members sharing similar CDR3s in the serum. In the heatmap at right, the edit distance between all pairs of serum CDR3s is shown. Two large families containing 9 or more members appear as blue sub matrices in the plot.
CDR3s with similar sequences may have similar specificity, and can be mined for additional hits. Every sequence recovered by Alicanto®, including those not selected as candidates, are delivered to you.
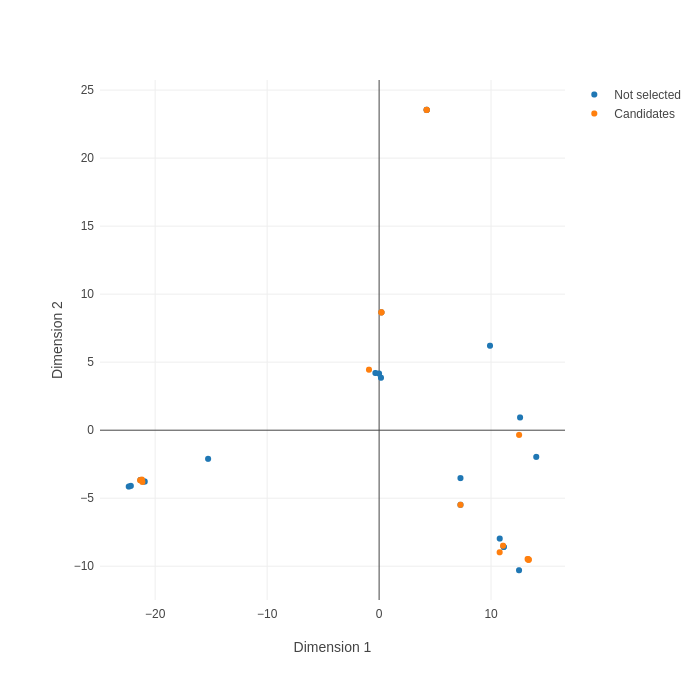
Alicanto® antibodies have diverse CDR3s
CDR3 sequence drives specificity and antibodies with similar CDR3 sequences are therefore likely to bind the same epitope on the target. For many applications, discovering antibodies that target diverse epitopes is desirable. In Alicanto®, we visualize the diversity of CDR3 sequences using multidimensional scaling (MDS).
In the MDS plot on the right, the edit distance of the CDR3s is used to generate the plot. Each point represents a distinct CDR3 identified by Alicanto®. Similar CDR3s appear close together in the plot, while CDR3s that have little sequence similarity are distant.
A final set of 15 CDR3s (shown in orange) were selected to ensure sequence diversity and maximize coverage of distinct epitopes.
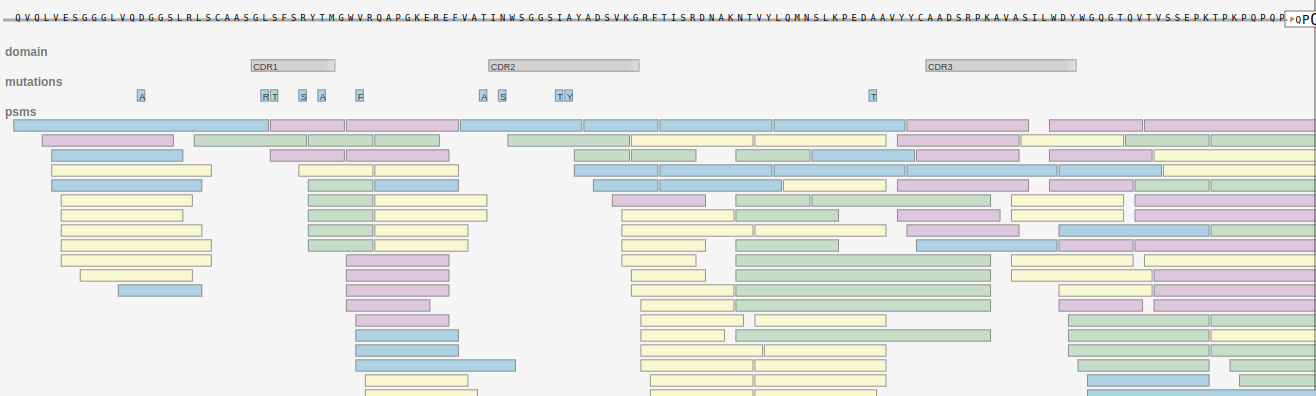
Alicanto® antibody candidate viewer
In the Alicanto® candidate viewer below a candidate, the sequence is shown with tracks denoting CDRs and mutations from the germline. Colored bars represent peptides derived from mass spectrometry that support the candidate sequence. The different colors indicate different enzymatic digests used to analyze the antibodies.
This candidate has strong evidence: 100% coverage of the sequence by peptides and evidence of affinity maturation in the form of mutations in the CDRs. Features of each candidate are also shown in the report including:
- Antibody Coverage – The percentage of the antibody variable region that is covered by peptides derived from mass spectrometry.
- CDR3 Coverage – The percentage of the CDR3 region that is covered by peptides derived from mass spectrometry.
- Clone Size – The number of distinct antibody sequences that are part of the same clone cluster as the candidate sequence and have highly similar CDR3 sequences.
- Mutation Count – The number of amino acid mutations incurred since germline V, D, J gene rearrangement.
- CDR Sequences – The amino acid sequences for CDRs 1, 2, and 3.
Related Resources
With Alicanto® you see not just the diverse candidates selected by our platform, but any relative of the candidates found in the repertoire. We recently wrote about Next Generation Sequencing-Enhanced Antibody Discovery on how using the full repertoire helps your antibody discovery or engineering project.
The Antibody Reference Guide provides information on antibodies including background information on isotypes, germine rearrangement, and antibody structure.