Immune Repertoire Sequencing in Human
Immune repertoire sequencing enables deeper understanding of the adaptive immune response [1]. Abterra Biosciences has developed Reptor™, a flexible immune repertoire sequencing and analysis service.
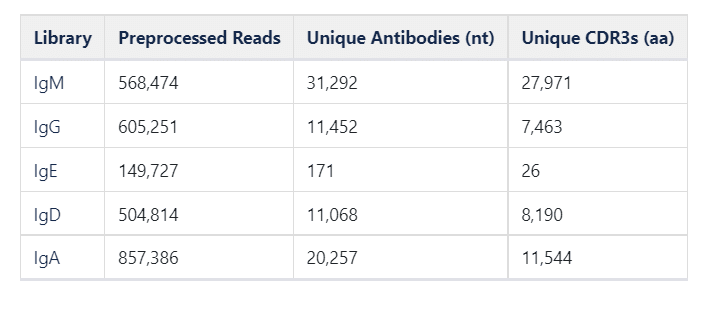
In this study, we analyzed the B-cell receptor (BCR) repertoire from peripheral blood mononuclear cells (PBMCs) from a healthy patient using Reptor, and examined repertoire characteristics across different heavy chain isotypes: IgG, IgM, IgA, IgD, and IgE.
Immune Repertoire Sequencing Sample and Data
From the PBMC sample, we extracted RNA, constructed cDNA, and aliquoted cDNA into 5 samples. From each sample, proprietary isotype-specific primers were used to amplify the variable region of BCRs in a 5′ RACE strategy. Libraries were loaded onto the Illumina MiSeq system and sampled to a depth of between 500K -1.5M 2×300 bp reads per library.
The sequencing reads are preprocessed by Reptor to remove non-antibody contaminants, low quality reads, and paired-end reads that cannot be stitched into a single sequence.
The MiSeq system has a documented error rate of 1%, virtually guaranteeing that every read will have at least one error. We previously wrote about the importance of error correction in immune repertoire sequencing, and how we’ve addressed this challenge in Reptor. After error correction, Reptor filters sequences that are below a minimum abundance threshold for error correction (2 reads), are too short or too long, or do not align well to germline gene segments.
Tell Us About Your Project
IgM expressing antibodies are more diverse than other isotypes
IgM isotypes are expressed in naïve B cells, as well as some memory B cells. Not unexpectedly, we see the greatest diversity among IgM antibodies. Compared to the other isotypes, IgM antibodies both have a larger number of total unique antibody sequences, the largest number of CDR3 sequences, and the lowest mean read count per antibody. 45% of the IgM antibodies had no mutations in the V gene segment compared to germline.
Clonal diversity of each isotype is measured as a function of empirical resampling of sequencing reads in the above rarefaction curve. The plot is automatically generated by Reptor, and provides a comparison of CDR3 diversity across the different isotypes. In addition, we see the curves have saturated and no additional CDR3s would be recovered by sequencing this library more deeply. The IgM isotype shows the greatest diversity in CDR3s, while IgE shows the least diversity. IgE expressed antibodies are the least diverse. IgE antibodies are involved in allergic reactions and immune responses to certain pathogens, but have been observed to be rare in blood, both in serum IgE abundance as well as IgE-expressing B cell frequency [2]. Because of the small size of the IgE repertoire, it was excluded from subsequent repertoire analysis.
Mutation distribution differs across isotypes
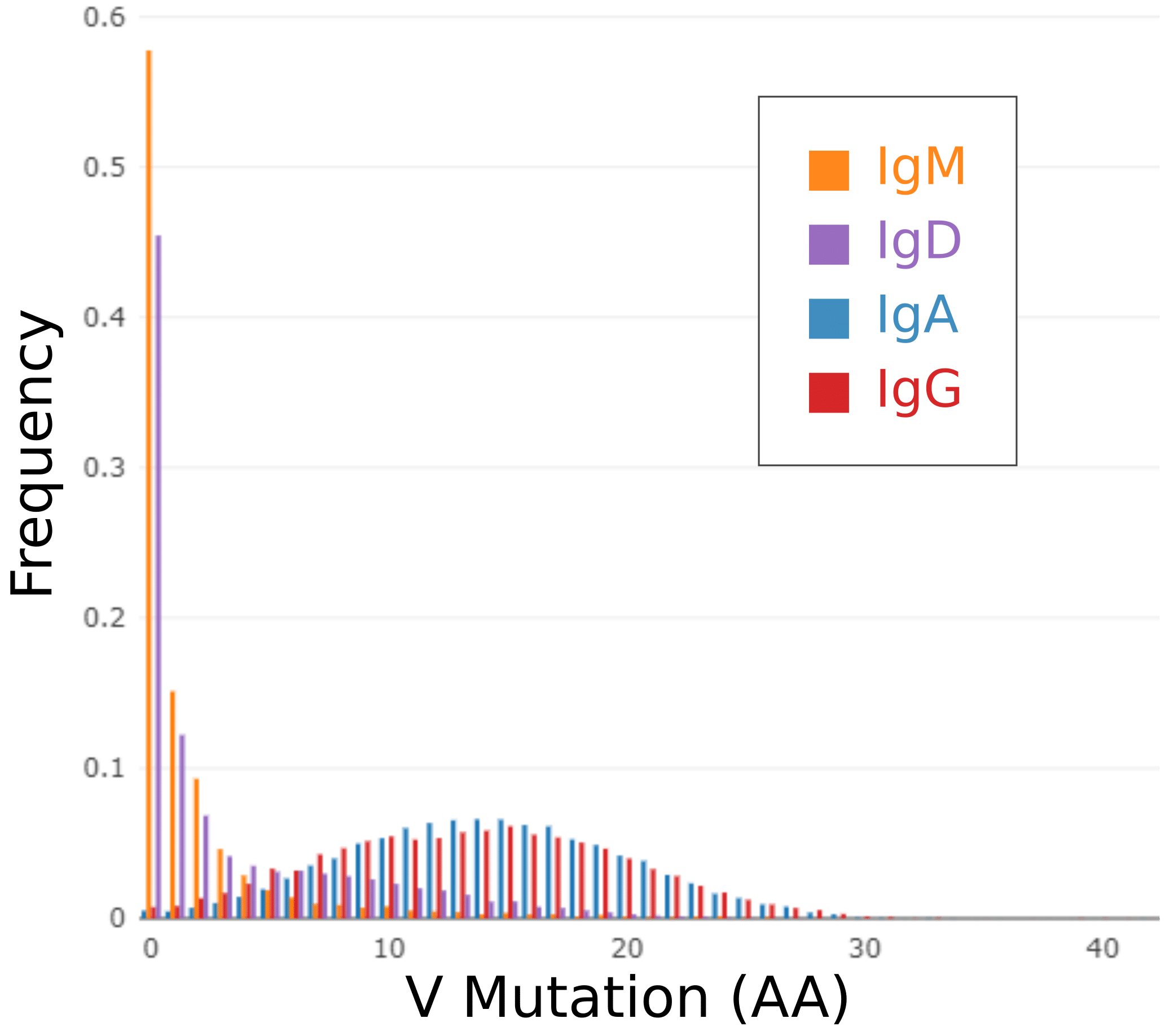
For each antibody, Reptor identifies mutations in the variable region with respect to the V gene germline. On the left, a standard Reptor output plot shows a histograph of frequency of antibodies versus amino acid mutation count. Each isotype is shown as a distinct series, and it becomes immediately clear that the repertoires can be categorized as either being predominantly naïve (IgD and IgM) or predominantly mutated (IgG and IgA).
CDR3 length is consistent across isotypes
A comparison of the CDR3 lengths across isotypes shows no marked difference between the repertoires. The mode for all isotypes was 14 amino acids, which is consistent with other documented repertoires [3].
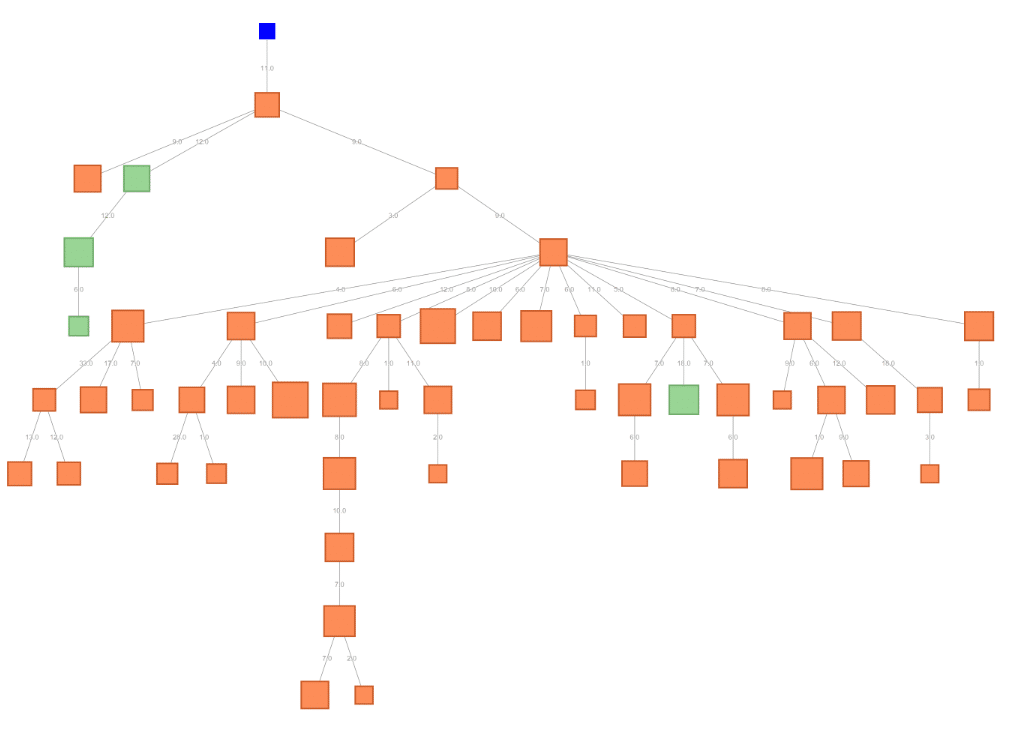
Few clones are expressed as multiple isotypes
Very few CDR3s were found in multiple isotypes, which is not unexpected in bulk sampling of BCRs from PBMCs. The greatest fraction of shared clones occurred between IgA and IgG where 3% of clones (569 of 18,438 CDR3s) in either repertoire were found in both repertoires. The IgM repertoire shared clones with both IgA (311 of 39,204 clones) and IgD (270 of 35,891 clones), but only 106 of 35,328 clones were shared with IgG.
Below, a minimum spanning tree on edit distance shows the relationship between different antibody sequences that share the same CDR3 sequence. Green boxes are from the IgG repertoire while orange boxes are from the IgA repertoire. The blue node is the theoretical (but not observed) germline root of this lineage.
Conclusion
We used Reptor, our flexible immune repertoire sequencing technology, to analyze the baseline BCR repertoire in a healthy human. From the analysis we were able to observe similarities and differences across isotypes. Reptor can be used for a wide range of applications including:
- Time-series analysis: Reptor identifies clones that persist across time, are arise in response to an immune challenge such as a vaccine.
- Spatial analysis: Reptor compares immune repertoires across tissue such a peripheral blood, lymph node, and bone marrow.
- Case-control analysis: Multiple patients can be analyzed by Reptor simultaneously to identify differences between classes.
Related Resources
[1] Georgiou, George, et al. “The promise and challenge of high-throughput sequencing of the antibody repertoire.” Nature biotechnology 32.2 (2014): 158-168. (PubMed)
[2] Levin, Mattias, et al. “Antibody-encoding repertoires of bone marrow and peripheral blood—a focus on IgE.” Journal of Allergy and Clinical Immunology 139.3 (2017): 1026-1030. (PubMed)
[3] Arnaout, Ramy, et al. “High-resolution description of antibody heavy-chain repertoires in humans.” PloS one 6.8 (2011): e22365. (PubMed)