From RNA to Sequencing: QC Matters
Sequencing the immunoglobulin (Ig) repertoire provides us with important information on the adaptive immune response, and can help with the development of diagnostic and therapeutic applications. A single human can have an estimated 10¹¹ B cells with millions of distinct Ig clonal populations. Sequencing even a fraction of this diversity has been practically and financially unrealistic until the advent of next generation sequencing (NGS). Sequencing the breadth of diversity of the Ig repertoire is a key component of our Alicanto® service. Through the course of sequencing Ig repertoires from a host of different species, we’ve discovered that QC matters. We’ve distilled some lessons into this post. We published a second post on constructing repertoires from reads.
While NGS allows scientists to survey DNA or RNA at the genomics/transcriptomics level, sequencing of the Ig repertoire (Ig-seq) comes with several inherent problems due to the biases and errors introduced by the techniques necessary to prepared the DNA/RNA for sequencing. In order to sample only the Ig repertoire during sequencing, reverse transcription and/or amplification of the DNA/RNA is necessary. However, this process, combined with the sequencing itself, is imperfect and will introduce errors that have to be correct post-sequencing (Friedensohn et al 2017).
Another source of bias comes from the primers that are needed to perform the amplification of the antibody transcripts. In many cases, sets of degenerate primers are used to amplify the variable region of the antibody sequence, which can lead to bias in amplification of certain sequences, e. g. biasing the representation of transcripts with certain V-genes (Carlson et al 2013). One way to minimize amplification biases is to use the 5′ rapid amplification of cDNA ends (5′ RACE) (Frohman et al 1988). This method only requires a single gene specific primer, targeted at the 3’ end of the desired region of the transcript. In the case of Ig-seq this is ideal because the primer can be designed to target the constant region of the transcript, which is significantly less variable than the portion of the transcript going towards the 5’ end (aptly named variable region).
In 5’ RACE, cDNA synthesis proceeds until the 5′ end of the target mRNA is reached, and then one of several approaches can be used to add a known, unrelated primer sequence to the 3′ end of the cDNA strand, permitting subsequent PCR amplification. Because of this, RNA with good integrity is particularly important when using 5’RACE during library prep, since any transcript that matched the gene specific primer at the 3’ end, should have the matching adapter on the 5’ end, independent of the level of degradation of the transcript. This means that any transcripts that match the reverse gene specific primer will be amplified and could be incorporated into the library, including degraded antibody transcripts, containing only a portion of the sequence. Performing size selection will ameliorate this problem, but as you’ll see below, if RNA degradation is high, the results can be very bad, with most sequences being composed mostly of sequencing adapters.
We sequenced the antibody repertoires of samples with varying degrees of RNA degradation, showing a direct correlation between RNA quality, quality of sequencing output, and repertoire construction. The results below are a great visual representation of how RNA degradation can have a significant effect on how much of the repertoire is sampled. Libraries were sequences as paired end 2 x 300bp. Adapter content plot refers to read 1 of the paired reads.
Tell Us About Your Project
RNA Integrity
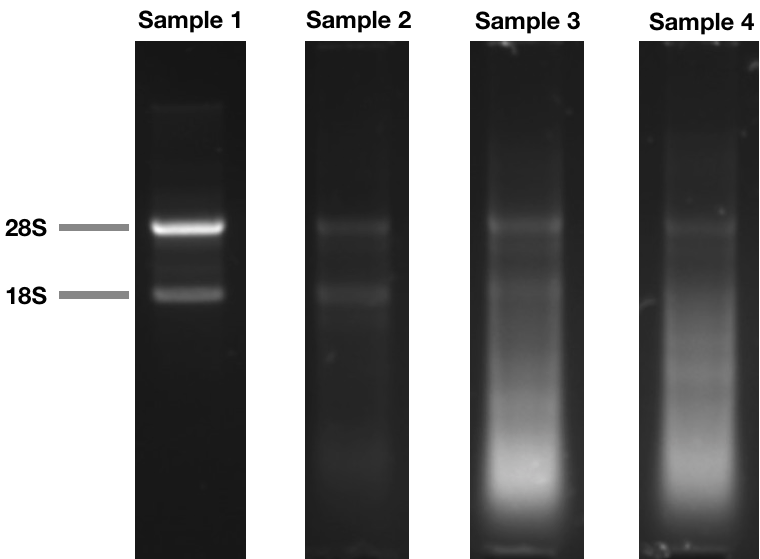
Agarose gel electrophoresis of RNA. One easy way to check for RNA integrity is to run an electrophoresis gel. RNA with good integrity will show two sharp bands (28S and 18S rRNA). The intensity of the 28S band should be twice as bright as the 18S band. As RNA degrades, the larger bands will become fainter, and a low molecular weight smear will appear. A denaturing gel is usually used to run RNA samples, but a non-denaturing agarose gel can also be used to visualize the 28S/18S bands. Other methods, requiring specialized equipment, can be used to calculate a RNA Integrity Number (RIN), which uses the 28S/18S ratio, or the Qubit RNA IQ, which measure the ratio between large and small RNA in the sample.
5′ RACE Amplicon
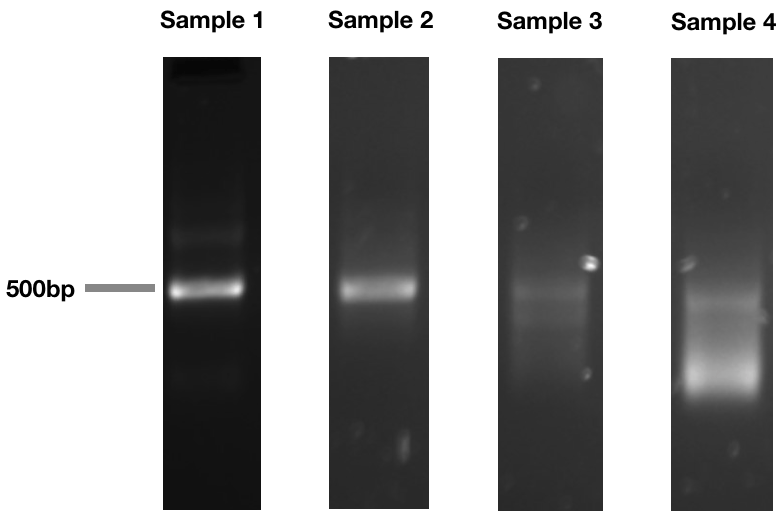
5’ RACE products for samples with increasing levels of RNA degradation. Notice how a sample with good RNA integrity has a sharp band, but as RNA integrity decreases, this band becomes a smear of low molecular weight.
Sequencing
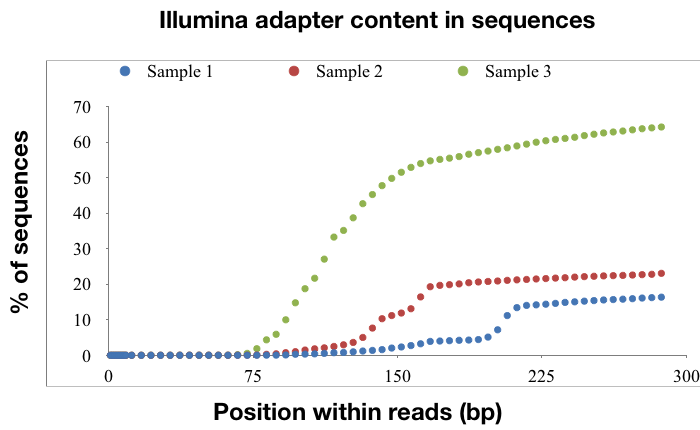
Illumina libraries were prepared with three of the samples and sequenced on the MiSeq as 2 x 300bp reads. QC analysis shows that the percentage of reads that are mostly composed of adapter sequence, instead of antibody sequence, increases drastically as RNA degradation increases. This was the case even though size selection to remove small amplicons from the library was performed. Note that we excluded sample 4 because the quality of the library was too low due to RNA degradation.
Repertoire construction
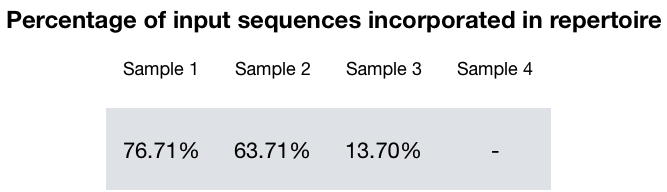
In order for a read to be included in the final Ig repertoire, it must pass several quality and content filters. The percentage of reads that pass all these filters decreases sharply as RNA integrity for the samples decreases. This means that with increasing RNA degradation, sampling of the Ig repertoire decreases significantly.
What to do to prevent RNA degradation?
The simplest method is to put cells or tissue in RNAlater as soon as possible. This is particularly important for tissues with high levels of RNases, such as spleen. However, this is not always possible, since RNAlater will denature proteins, which might prevent cell sorting. In these cases, keeping the cells alive and minimizing the time between the processing of cells and RNA extraction is important.