NGS During The Hybridoma Generation Process
Next-generation sequencing (NGS) has allowed for unprecedented insights into patterns of the immune response. It can enable high-throughput sequencing of hybridomas as we have discussed in NGS-Enhanced antibody discovery blog post and resource pages. NGS can provide a deeper understanding into the diversity of the immune response and clones that persist from mouse to hybridoma cell line. In this blog post, we will:
-
-
-
Demonstrate the use of NGS and our Reptor software throughout the hybridoma generation process including from remnant tissue, the hybridoma fusion pool, and expanded hybridoma clones.
-
Identify clonal lineages that gave rise to screened hybridoma hits.
-
-
Hybridoma Generation Background
If you are unfamiliar with the hybridoma generation process, please see our hybridoma resource pages. We immunized a cohort of 6 mice with a CD3 fragment. We collected lymph node (LN) and spleen (SP) from 2 of the mice, a portion of which was reserved for NGS analysis. The remaining tissue was fused with a myeloma cell line to produce a hybridoma fusion library (Hyb-Lib). Finally, the fusions were sorted into mono- and penta-clonal cultures and screened for reactivity to CD3. A final set of 18 monoclonal hybridomas (Hyb-Select) were selected for cell culture and sequencing with Reptor.
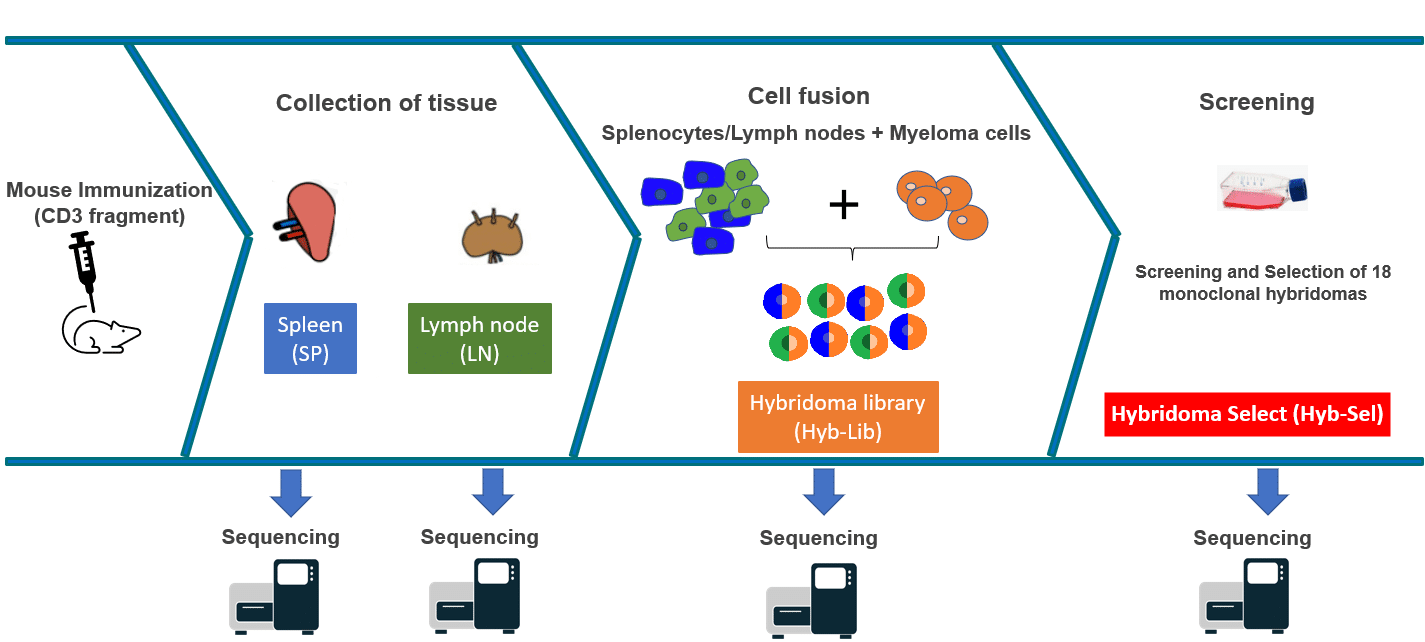
Reptor™ hybridoma sequencing process
Reptor™ is a flexible, NGS-powered service for B-cell receptor repertoire sequencing from RNA. For each of the four sources (LN, SP, Hyb-Lib, and Hyb-Select), Reptor was used to extract RNA, amplify the variable regions of the IgH and IgK/IgL transcripts, sequence the amplicons, and analyze the sequencing reads to construct a B-cell receptor repertoire.
Lymph nodes and Spleens
The lymph nodes (LN) and spleens (SP) from the immunized mice were pooled across mice prior to analysis. While these tissues contain a heterogeneous mix of cell types, Reptor amplifies the B-cell receptor sequences to avoid sequencing non-B-cell receptor transcripts. IgG and IgM isotype transcripts were analyzed for the heavy chain. Approximately 1M 2×300 bp reads were generated for each source tissue and each chain.
Hybridoma Library
The hybridoma library (Hyb-Lib) is the intermediate product after hybridoma fusion but before sub-cloning. It captures the diversity of the immune response that can be subsequently recovered in monoclonal or oligoclonal hybridomas. It is presumed to be less diverse than the source tissue but more diverse than the screened hybridomas; where it lies on that spectrum could be project and hybridoma generation process specific. In this project, the Hyb-Lib was analyzed in the same manner as the standard tissue; however, unlike LN and SP all input cells should be producing a functional antibody transcript. Approximately 1M 2×300 bp reads were generated for each chain.
Clone diversity during hybridoma generation
The three bulk samples were analyzed starting from similar amounts of RNA and sequenced to similar depth (~1M reads). Interestingly, the LN sample appeared the most diverse in terms of unique CDR3 sequence, followed by SP. The Hyb-Lib had the smallest number of distinct CDR3 sequences. We define a clone by it’s amino acid CDR3 sequence, so any sequences that share the same CDR3 are part of the same clone. A clone’s size is equivalent to the number of sequences sharing the same CDR3.
The LN sample had both the largest clone (590 antibodies sharing the same CDR3) and the smallest mean clone size (3 sequences), suggesting the distribution of clone sizes has a very long tail of very small clones.
Clone overlap between hybridoma generation stages
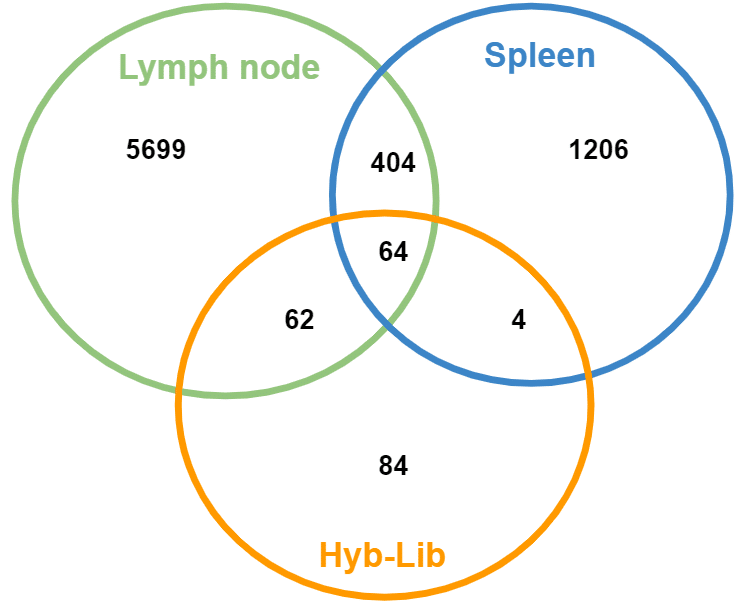
Comparing co-occurrence of clones across the three samples there are several interesting observations:
-
-
-
Lymph node (LN) and spleen (SP) are fairly distinct with a Jaccard similarity of 0.06. This is consistent with clonal overlap across tissues in other immunization projects. In past projects, technical replicates from the same individual can have Jaccard similarity of more than 0.4, while samplings from distinct individuals have near 0 overlap for most species.
-
The Hyb-Lib has more in common with the LN than SP; however, nearly 40% of the Hyb-Lib clones are unique to that sample.
-
A minority of clones discovered in the mouse tissue were identified in the Hyb-Lib (130/7439).
-
-
Our 18 selected hybridomas resulted in 11 unique CDR3s. Six of these were shared across all samples, 3 appeared only in the Hyb-Lib, 1 appeared in Hyb-Lib and LN, and 1 appeared in none of the samples.
Next steps: Hit expansion in hybridoma generation
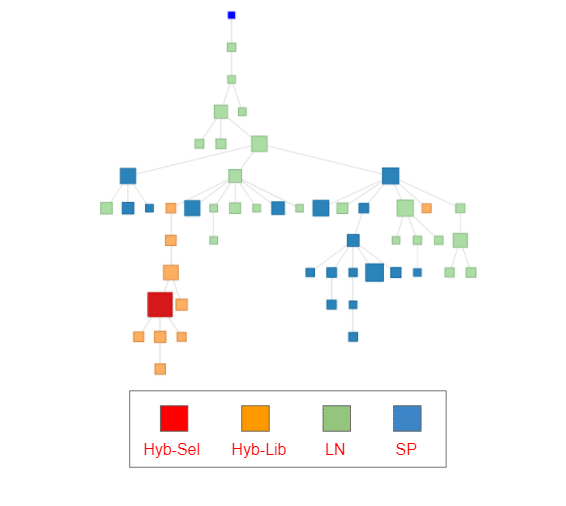
NGS reveals relatives of screened hits that can be used for hit expansion, as we’ve described in a previous blog post for llama single domain antibody discovery. In the figure to the left, we constructed a minimum spanning tree (MST) on antibody sequences from the same clone as one of our 11 selected hybridoma sequences.
The MST is useful for visualizing similarity between sequences but should not be used to infer phylogenetic relationships. This particular clone appears across all 3 bulk samples (LN, SP, Hyb-Lib) and contains sequences that are of varying level of mutation to the selected hit (Red). The size of the blocks indicates the read count.
Get In Touch